Solvent-induced differentiation of protein backbone hydrogen bonds in calmodulin
- PMID: 17567747
- PMCID: PMC2206704
- DOI: 10.1110/ps.062689807
Solvent-induced differentiation of protein backbone hydrogen bonds in calmodulin
Abstract
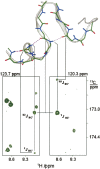
In apo and holoCaM, almost half of the hydrogen bonds (H-bonds) at the protein backbone expected from the corresponding NMR or X-ray structures were not detected by h3JNC' couplings. The paucity of the h3JNC' couplings was considered in terms of dynamic features of these structures. We examined a set of seven proteins and found that protein-backbone H-bonds form two groups according to the h3JNC' couplings measured in solution. H-bonds that have h3JNC' couplings above the threshold of 0.2 Hz show distance/angle correlation among the H-bond geometrical parameters, and appear to be supported by the backbone dynamics in solution. The other H-bonds have no such correlation; they populate the water-exposed and flexible regions of proteins, including many of the CaM helices. The observed differentiation in a dynamical behavior of backbone H-bonds in apo and holoCaM appears to be related to protein functions.
Figures






Comment in
-
NMR provides evidence for dynamic hydrogen bonding in proteins.Protein Sci. 2007 Jul;16(7):1247-8. doi: 10.1110/ps.072945407. Epub 2007 Jun 13. Protein Sci. 2007. PMID: 17567735 Free PMC article. No abstract available.
References
-
- Assadi-Porter F.M., Abildgaard, F., Blad, H., and Markley, J.L. 2003. Correlation of the sweetness of variants of the protein brazzein with patterns of hydrogen bonds detected by NMR spectroscopy. J. Biol. Chem. 278: 31331–31339. - PubMed
-
- Barabato G., Ikura, M., Kay, L.E., Pastor, R.W., and Bax, A. 1992. Backbone dynamics of calmodulin studied by 15N relaxation using inverse detected two-dimensional NMR spectroscopy: The central helix is flexible. Biochemistry 31: 5269–5278. - PubMed
-
- Barfield M. 2002. Structural dependencies of interresidue scalar coupling h3 J NC, and donor 1H chemical shifts in the hydrogen bonding regions of proteins. J. Am. Chem. Soc. 124: 4158–4168. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

