A domain-based approach to predict protein-protein interactions
- PMID: 17567909
- PMCID: PMC1919395
- DOI: 10.1186/1471-2105-8-199
A domain-based approach to predict protein-protein interactions
Abstract
Background: Knowing which proteins exist in a certain organism or cell type and how these proteins interact with each other are necessary for the understanding of biological processes at the whole cell level. The determination of the protein-protein interaction (PPI) networks has been the subject of extensive research. Despite the development of reasonably successful methods, serious technical difficulties still exist. In this paper we present DomainGA, a quantitative computational approach that uses the information about the domain-domain interactions to predict the interactions between proteins.
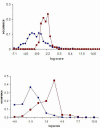
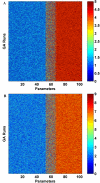
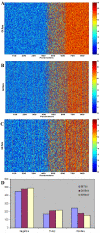
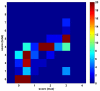
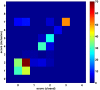
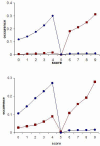
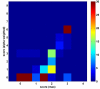
Results: DomainGA is a multi-parameter optimization method in which the available PPI information is used to derive a quantitative scoring scheme for the domain-domain pairs. Obtained domain interaction scores are then used to predict whether a pair of proteins interacts. Using the yeast PPI data and a series of tests, we show the robustness and insensitivity of the DomainGA method to the selection of the parameter sets, score ranges, and detection rules. Our DomainGA method achieves very high explanation ratios for the positive and negative PPIs in yeast. Based on our cross-verification tests on human PPIs, comparison of the optimized scores with the structurally observed domain interactions obtained from the iPFAM database, and sensitivity and specificity analysis; we conclude that our DomainGA method shows great promise to be applicable across multiple organisms.
Conclusion: We envision the DomainGA as a first step of a multiple tier approach to constructing organism specific PPIs. As it is based on fundamental structural information, the DomainGA approach can be used to create potential PPIs and the accuracy of the constructed interaction template can be further improved using complementary methods. Explanation ratios obtained in the reported test case studies clearly show that the false prediction rates of the template networks constructed using the DomainGA scores are reasonably low, and the erroneous predictions can be filtered further using supplementary approaches such as those based on literature search or other prediction methods.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

