Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia
- PMID: 17568569
- PMCID: PMC2080683
- DOI: 10.1016/j.biopsych.2006.12.021
Molecular evidence for increased expression of genes related to immune and chaperone function in the prefrontal cortex in schizophrenia
Abstract
Background: Schizophrenia is characterized by complex gene expression changes. The transcriptome alterations in the prefrontal cortex have been the subject of several recent postmortem studies that yielded both convergent and divergent findings.
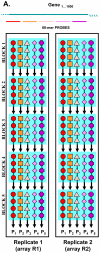
Methods: To increase measurement precision, we used a custom-designed DNA microarray platform with long oligonucleotides and multiple probes with replicates. The platform was designed to assess the expression of > 1800 genes specifically chosen because of their hypothesized roles in the pathophysiology of schizophrenia. The gene expression differences in dorsolateral prefrontal cortex samples from 14 matched pairs of schizophrenia and control subjects were analyzed with two technical replicates and four data mining approaches.
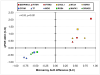
Results: In addition to replicating many expression changes in synaptic, oligodendrocyte, and signal transduction genes, we uncovered and validated a robust immune/chaperone transcript upregulation in the schizophrenia samples.
Conclusions: We speculate that the overexpression of SERPINA3, IFITM1, IFITM2, IFITM3, CHI3L1, MT2A, CD14, HSPB1, HSPA1B, and HSPA1A in schizophrenia subjects represents a long-lasting and correlated signature of an early environmental insult during development that actively contributes to the pathophysiology of prefrontal dysfunction.
Figures



References
-
- Altar CA, Jurata LW, Charles V, et al. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. - PubMed
-
- Ashdown H, Dumont Y, Ng M, Poole S, Boksa P, Luheshi GN. The role of cytokines in mediating effects of prenatal infection on the fetus: implications for schizophrenia. Mol Psychiatry. 2006;11:47–55. - PubMed
-
- Aston C, Jiang L, Sokolov BP. Microarray analysis of postmortem temporal cortex from patients with schizophrenia. J Neurosci Res. 2004;77:858–66. - PubMed
-
- Cannon M, Clarke MC. Risk for schizophrenia--broadening the concepts, pushing back the boundaries. Schizophr Res. 2005;79:5–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

