SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK
- PMID: 17568778
- PMCID: PMC1914091
- DOI: 10.1038/sj.emboj.7601743
SINTBAD, a novel component of innate antiviral immunity, shares a TBK1-binding domain with NAP1 and TANK
Abstract
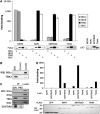
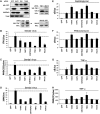
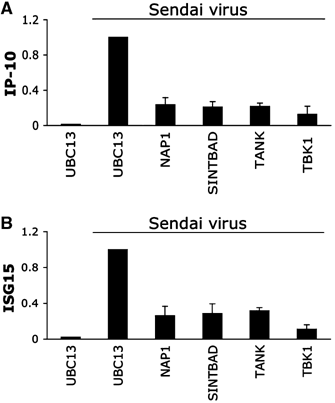
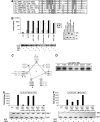
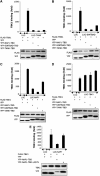
The expression of antiviral genes during infection is controlled by inducible transcription factors such as IRF3 (interferon regulatory factor). Activation of IRF3 requires its phosphorylation by TBK1 (TANK-binding kinase) or IKKi (inhibitor of nuclear factor kappaB kinase, inducible). We have identified a new and essential component of this pathway, the adaptor protein SINTBAD (similar to NAP1 TBK1 adaptor). SINTBAD constitutively binds TBK1 and IKKi but not related kinases. Upon infection with Sendai virus, SINTBAD is essential for the efficient induction of IRF-dependent transcription, as are two further TBK1 adaptors, TANK and NAP1. We identified a conserved TBK1/IKKi-binding domain (TBD) in the three adaptors, predicted to form an alpha-helix with residues essential for kinase binding clustering on one side. Isolated TBDs compete with adaptor binding to TBK1 and prevent poly(I:C)-induced IRF-dependent transcription. Our results suggest that efficient signal transduction upon viral infection requires SINTBAD, TANK and NAP1 because they link TBK1 and IKKi to virus-activated signalling cascades.
Figures


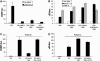
Similar articles
-
Functional dissection of the TBK1 molecular network.PLoS One. 2011;6(9):e23971. doi: 10.1371/journal.pone.0023971. Epub 2011 Sep 8. PLoS One. 2011. PMID: 21931631 Free PMC article.
-
TRIM11 negatively regulates IFNβ production and antiviral activity by targeting TBK1.PLoS One. 2013 May 13;8(5):e63255. doi: 10.1371/journal.pone.0063255. Print 2013. PLoS One. 2013. PMID: 23675467 Free PMC article.
-
MAVS activates TBK1 and IKKε through TRAFs in NEMO dependent and independent manner.PLoS Pathog. 2017 Nov 10;13(11):e1006720. doi: 10.1371/journal.ppat.1006720. eCollection 2017 Nov. PLoS Pathog. 2017. PMID: 29125880 Free PMC article.
-
Adaptor-Mediated Trafficking of Tank Binding Kinase 1 During Diverse Cellular Processes.Traffic. 2025 Jan-Mar;26(1-3):e70000. doi: 10.1111/tra.70000. Traffic. 2025. PMID: 40047067 Free PMC article. Review.
-
Regulation of IRF3 activation in human antiviral signaling pathways.Biochem Pharmacol. 2022 Jun;200:115026. doi: 10.1016/j.bcp.2022.115026. Epub 2022 Mar 31. Biochem Pharmacol. 2022. PMID: 35367198 Review.
Cited by
-
The role of immunostimulatory nucleic acids in septic shock.Int J Clin Exp Med. 2012;5(1):1-23. Epub 2012 Jan 15. Int J Clin Exp Med. 2012. PMID: 22328944 Free PMC article.
-
The IKK Kinases: Operators of Antiviral Signaling.Viruses. 2010 Jan;2(1):55-72. doi: 10.3390/v2010055. Epub 2010 Jan 8. Viruses. 2010. PMID: 21994600 Free PMC article.
-
Use of the pharmacological inhibitor BX795 to study the regulation and physiological roles of TBK1 and IkappaB kinase epsilon: a distinct upstream kinase mediates Ser-172 phosphorylation and activation.J Biol Chem. 2009 May 22;284(21):14136-46. doi: 10.1074/jbc.M109.000414. Epub 2009 Mar 22. J Biol Chem. 2009. PMID: 19307177 Free PMC article.
-
A multi-step genomic approach prioritized TBKBP1 gene as relevant for multiple sclerosis susceptibility.J Neurol. 2022 Aug;269(8):4510-4522. doi: 10.1007/s00415-022-11109-8. Epub 2022 May 12. J Neurol. 2022. PMID: 35545683 Free PMC article.
-
The tyrosine kinase c-Src enhances RIG-I (retinoic acid-inducible gene I)-elicited antiviral signaling.J Biol Chem. 2009 Jul 10;284(28):19122-31. doi: 10.1074/jbc.M808233200. Epub 2009 May 6. J Biol Chem. 2009. PMID: 19419966 Free PMC article.
References
-
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA (2001) Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature 413: 732–738 - PubMed
-
- Barrios-Rodiles M, Brown KR, Ozdamar B, Bose R, Liu Z, Donovan RS, Shinjo F, Liu Y, Dembowy J, Taylor IW, Luga V, Przulj N, Robinson M, Suzuki H, Hayashizaki Y, Jurisica I, Wrana JL (2005) High-throughput mapping of a dynamic signaling network in mammalian cells. Science 307: 1621–1625 - PubMed
-
- Bouwmeester T, Bauch A, Ruffner H, Angrand PO, Bergamini G, Croughton K, Cruciat C, Eberhard D, Gagneur J, Ghidelli S, Hopf C, Huhse B, Mangano R, Michon AM, Schirle M, Schlegl J, Schwab M, Stein MA, Bauer A, Casari G, Drewes G, Gavin AC, Jackson DB, Joberty G, Neubauer G, Rick J, Kuster B, Superti-Furga G (2004) A physical and functional map of the human TNF-alpha/NF-kappa B signal transduction pathway. Nat Cell Biol 6: 97–105 - PubMed
-
- Connelly MA, Marcu KB (1995) CHUK, a new member of the helix-loop-helix and leucine zipper families of interacting proteins, contains a serine-threonine kinase catalytic domain. Cell Mol Biol Res 41: 537–549 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

