Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis
- PMID: 17569673
- PMCID: PMC1919493
- DOI: 10.1093/nar/gkm388
Functional analysis of Saccharomyces cerevisiae ribosomal protein Rpl3p in ribosome synthesis
Abstract
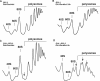
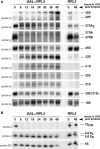
Ribosome synthesis in eukaryotes requires a multitude of trans-acting factors. These factors act at many steps as the pre-ribosomal particles travel from the nucleolus to the cytoplasm. In contrast to the well-studied trans-acting factors, little is known about the contribution of the ribosomal proteins to ribosome biogenesis. Herein, we have analysed the role of ribosomal protein Rpl3p in 60S ribosomal subunit biogenesis. In vivo depletion of Rpl3p results in a deficit in 60S ribosomal subunits and the appearance of half-mer polysomes. This phenotype is likely due to the instability of early and intermediate pre-ribosomal particles, as evidenced by the low steady-state levels of 27SA(3), 27SB(S) and 7S(L/S) precursors. Furthermore, depletion of Rpl3p impairs the nucleocytoplasmic export of pre-60S ribosomal particles. Interestingly, flow cytometry analysis indicates that Rpl3p-depleted cells arrest in the G1 phase. Altogether, we suggest that upon depletion of Rpl3p, early assembly of 60S ribosomal subunits is aborted and subsequent steps during their maturation and export prevented.
Figures




Similar articles
-
Role of the yeast ribosomal protein L16 in ribosome biogenesis.FEBS J. 2016 Aug;283(16):2968-85. doi: 10.1111/febs.13797. Epub 2016 Jul 15. FEBS J. 2016. PMID: 27374275
-
Ribosomal protein L35 is required for 27SB pre-rRNA processing in Saccharomyces cerevisiae.Nucleic Acids Res. 2010 Aug;38(15):5177-92. doi: 10.1093/nar/gkq260. Epub 2010 Apr 14. Nucleic Acids Res. 2010. PMID: 20392820 Free PMC article.
-
The essential WD-repeat protein Rsa4p is required for rRNA processing and intra-nuclear transport of 60S ribosomal subunits.Nucleic Acids Res. 2005 Oct 12;33(18):5728-39. doi: 10.1093/nar/gki887. Print 2005. Nucleic Acids Res. 2005. PMID: 16221974 Free PMC article.
-
Nuclear export and cytoplasmic maturation of ribosomal subunits.FEBS Lett. 2007 Jun 19;581(15):2783-93. doi: 10.1016/j.febslet.2007.05.013. Epub 2007 May 11. FEBS Lett. 2007. PMID: 17509569 Review.
-
Pre-ribosomes on the road from the nucleolus to the cytoplasm.Trends Cell Biol. 2003 May;13(5):255-63. doi: 10.1016/s0962-8924(03)00054-0. Trends Cell Biol. 2003. PMID: 12742169 Review.
Cited by
-
Las1L is a nucleolar protein required for cell proliferation and ribosome biogenesis.Mol Cell Biol. 2010 Sep;30(18):4404-14. doi: 10.1128/MCB.00358-10. Epub 2010 Jul 20. Mol Cell Biol. 2010. PMID: 20647540 Free PMC article.
-
Studies on the Coordination of Ribosomal Protein Assembly Events Involved in Processing and Stabilization of Yeast Early Large Ribosomal Subunit Precursors.PLoS One. 2015 Dec 7;10(12):e0143768. doi: 10.1371/journal.pone.0143768. eCollection 2015. PLoS One. 2015. PMID: 26642313 Free PMC article.
-
Feedback regulation of ribosome assembly.Curr Genet. 2018 Apr;64(2):393-404. doi: 10.1007/s00294-017-0764-x. Epub 2017 Oct 11. Curr Genet. 2018. PMID: 29022131 Review.
-
Rbp95 binds to 25S rRNA helix H95 and cooperates with the Npa1 complex during early pre-60S particle maturation.Nucleic Acids Res. 2022 Sep 23;50(17):10053-10077. doi: 10.1093/nar/gkac724. Nucleic Acids Res. 2022. PMID: 36018804 Free PMC article.
-
H/ACA snoRNA levels are regulated during stem cell differentiation.Nucleic Acids Res. 2020 Sep 4;48(15):8686-8703. doi: 10.1093/nar/gkaa612. Nucleic Acids Res. 2020. PMID: 32710630 Free PMC article.
References
-
- Olson MO, Dundr M, Szebeni A. The nucleolus: an old factory with unexpected capabilities. Trends Cell Biol. 2000;10:189–196. - PubMed
-
- Udem SA, Warner JR. The cytoplasmic maturation of a ribosomal precursor ribonucleic acid in yeast. J. Biol. Chem. 1973;248:1412–1416. - PubMed
-
- Trapman J, Retèl J, Planta RJ. Ribosomal precursor particles from yeast. Exp. Cell Res. 1975;90:95–104. - PubMed
-
- Takahashi N, Yanagida M, Fujiyama S, Hayano T, Isobe T. Proteomic snapshot analyses of preribosomal ribonucleoprotein complexes formed at various stages of ribosome biogenesis in yeast and mammalian cells. Mass Spectrom. Rev. 2003;22:287–317. - PubMed
-
- de la Cruz J, Kressler D, Linder P. In: Nucleolus. Olson MOJ, editor. Landes Bioscience/eurekah.com, Georgetown: Kluwer Academic; 2004. pp. 258–285.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

