Canalization of segmentation and its evolution in Drosophila
- PMID: 17569783
- PMCID: PMC1891814
- DOI: 10.1073/pnas.0701359104
Canalization of segmentation and its evolution in Drosophila
Abstract
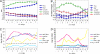
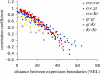
Segmentation in Drosophila embryogenesis occurs through a hierarchical cascade of regulatory gene expression driven by the establishment of a diffusion-mediated morphogen gradient. Here, we investigate the response of this pattern formation process to genetic variation and evolution in egg size. Specifically, we ask whether spatial localization of gap genes Kruppel (Kr) and giant (gt) and the pair-rule gene even-skipped (eve) during cellularization is robust to genetic variation in embryo length in three Drosophila melanogaster isolines and two closely related species. We identified two wild-derived strains of D. melanogaster whose eggs differ by approximately 25% in length when reared under identical conditions. These two lines, a D. melanogaster laboratory stock (w1118), and offspring from crosses between the lines all exhibit precise scaling in the placement of gap and pair-rule gene expression along the anterior-posterior axis in relation to embryo length. Genetic analysis indicates that this scaling is maternally controlled. Maternal regulation of scaling must be required for consistent localization of segmentation gene expression because embryo size, a genetically variable and adaptive trait, is maternally inherited. We also investigated spatial scaling between these D. melanogaster lines and single lines of Drosophila sechellia and Drosophila simulans, the latter two differing by approximately 25% in egg length. In contrast to the robust scaling we observed within species, localization of gene expression relative to embryo length differs significantly between the three species. Thus, the developmental mechanism that assures robust scaling within a species does not prevent rapid evolution between species.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

