Using structural motif descriptors for sequence-based binding site prediction
- PMID: 17570148
- PMCID: PMC1892084
- DOI: 10.1186/1471-2105-8-S4-S5
Using structural motif descriptors for sequence-based binding site prediction
Abstract
Background: Many protein sequences are still poorly annotated. Functional characterization of a protein is often improved by the identification of its interaction partners. Here, we aim to predict protein-protein interactions (PPI) and protein-ligand interactions (PLI) on sequence level using 3D information. To this end, we use machine learning to compile sequential segments that constitute structural features of an interaction site into one profile Hidden Markov Model descriptor. The resulting collection of descriptors can be used to screen sequence databases in order to predict functional sites.
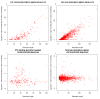
Results: We generate descriptors for 740 classified types of protein-protein binding sites and for more than 3,000 protein-ligand binding sites. Cross validation reveals that two thirds of the PPI descriptors are sufficiently conserved and significant enough to be used for binding site recognition. We further validate 230 PPIs that were extracted from the literature, where we additionally identify the interface residues. Finally we test ligand-binding descriptors for the case of ATP. From sequences with Swiss-Prot annotation "ATP-binding", we achieve a recall of 25% with a precision of 89%, whereas Prosite's P-loop motif recognizes an equal amount of hits at the expense of a much higher number of false positives (precision: 57%). Our method yields 771 hits with a precision of 96% that were not previously picked up by any Prosite-pattern.
Conclusion: The automatically generated descriptors are a useful complement to known Prosite/InterPro motifs. They serve to predict protein-protein as well as protein-ligand interactions along with their binding site residues for proteins where merely sequence information is available.
Figures



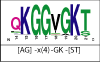

References
-
- Tong AHY, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M, Chen Y, Cheng X, Chua G, Friesen H, Goldberg DS, Haynes J, Humphries C, He G, Hussein S, Ke L, Krogan N, Li Z, Levinson JN, Lu H, Menard P, Munyana C, Parsons AB, Ryan O, Tonikian R, Roberts T, Sdicu AM, Shapiro J, Sheikh B, Suter B, Wong SL, Zhang LV, Zhu H, Burd CG, Munro S, Sander C, Rine J, Greenblatt J, Peter M, Bretscher A, Bell G, Roth FP, Brown GW, Andrews B, Bussey H, Boone C. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–813. doi: 10.1126/science.1091317. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

