Systematic discovery of in vivo phosphorylation networks
- PMID: 17570479
- PMCID: PMC2692296
- DOI: 10.1016/j.cell.2007.05.052
Systematic discovery of in vivo phosphorylation networks
Abstract
Protein kinases control cellular decision processes by phosphorylating specific substrates. Thousands of in vivo phosphorylation sites have been identified, mostly by proteome-wide mapping. However, systematically matching these sites to specific kinases is presently infeasible, due to limited specificity of consensus motifs, and the influence of contextual factors, such as protein scaffolds, localization, and expression, on cellular substrate specificity. We have developed an approach (NetworKIN) that augments motif-based predictions with the network context of kinases and phosphoproteins. The latter provides 60%-80% of the computational capability to assign in vivo substrate specificity. NetworKIN pinpoints kinases responsible for specific phosphorylations and yields a 2.5-fold improvement in the accuracy with which phosphorylation networks can be constructed. Applying this approach to DNA damage signaling, we show that 53BP1 and Rad50 are phosphorylated by CDK1 and ATM, respectively. We describe a scalable strategy to evaluate predictions, which suggests that BCLAF1 is a GSK-3 substrate.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
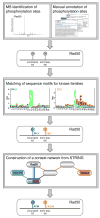
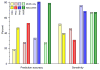
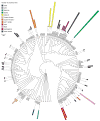
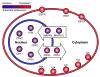



References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Aloy P, Russell RB. The third dimension for protein interactions and complexes. Trends Biochem Sci. 2002;27:633–8. - PubMed
-
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

