Celsius: a community resource for Affymetrix microarray data
- PMID: 17570842
- PMCID: PMC2394754
- DOI: 10.1186/gb-2007-8-6-r112
Celsius: a community resource for Affymetrix microarray data
Abstract
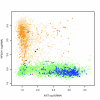
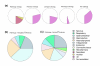
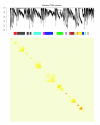
Celsius is a data warehousing system to aggregate Affymetrix CEL files and associated metadata. It provides mechanisms for importing, storing, querying, and exporting large volumes of primary and pre-processed microarray data. Celsius contains ten billion assay measurements and affiliated metadata. It is the largest publicly available source of Affymetrix microarray data, and through sheer volume it allows a sophisticated, broad view of transcription that has not previously been possible.
Figures




References
-
- Sarkans U, Parkinson H, Lara G, Oezcimen A, Sharma A, Abeygunawardena N, Contrino S, Holloway E, Rocca-Serra P, Mukherjee G, et al. The ArrayExpress gene expression database: a software engineering and implementation perspective. Bioinformatics. 2005;21:1495–1501. - PubMed
-
- Stuart J, Segal E, Koller D, Kim S. A gene-coexpression network for global discovery of conserved genetic modules. Science. 2003;302:249–255. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

