Virulence phenotypes of low-passage clinical isolates of nontypeable Haemophilus influenzae assessed using the chinchilla laniger model of otitis media
- PMID: 17570853
- PMCID: PMC1914350
- DOI: 10.1186/1471-2180-7-56
Virulence phenotypes of low-passage clinical isolates of nontypeable Haemophilus influenzae assessed using the chinchilla laniger model of otitis media
Abstract
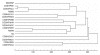
Background: The nontypeable Haemophilus influenzae (NTHi) are associated with a spectrum of respiratory mucosal infections including: acute otitis media (AOM); chronic otitis media with effusion (COME); otorrhea; locally invasive diseases such as mastoiditis; as well as a range of systemic disease states, suggesting a wide range of virulence phenotypes. Genomic studies have demonstrated that each clinical strain contains a unique genic distribution from a population-based supragenome, the distributed genome hypothesis. These diverse clinical and genotypic findings suggest that each NTHi strain possesses a unique set of virulence factors that contributes to the course of the disease.
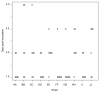
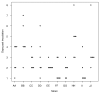
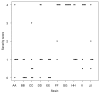
Results: The local and systemic virulence patterns of ten genomically characterized low-passage clinical NTHi strains (PittAA - PittJJ) obtained from children with COME or otorrhea were stratified using the chinchilla model of otitis media (OM). Each isolate was used to bilaterally inoculate six animals and thereafter clinical assessments were carried out daily for 8 days by blinded observers. There was no statistical difference in the time it took for any of the 10 NTHi strains to induce otologic (local) disease with respect to any or all of the other strains, however the differences in time to maximal local disease and the severity of local disease were both significant between the strains. Parameters of systemic disease indicated that the strains were not all equivalent: time to development of the systemic disease, maximal systemic scores and mortality were all statistically different among the strains. PittGG induced 100% mortality while PittBB, PittCC, and PittEE produced no mortality. Overall Pitt GG, PittII, and Pitt FF produced the most rapid and most severe local and systemic disease. A post hoc determination of the clinical origins of the 10 NTHi strains revealed that these three strains were of otorrheic origin, whereas the other 7 were from patients with COME.
Conclusion: Collectively these data suggest that the chinchilla OM model is useful for discriminating between otorrheic and COME NTHi strains as to their disease-producing potential in humans, and combined with whole genome analyses, point the way towards identifying classes of virulence genes.
Figures



References
-
- Murphy TF. Respiratory infections caused by non-typeable Haemophilus influenzae. Curr Opin Infect Dis. 2003;16:129–34. - PubMed
-
- Greenberg D, Broides A, Blancovich I, Peled N, Givon-Lavi N, Dagan R. Relative importance of nasopharyngeal versus oropharyngeal sampling for isolation of Streptococcus pneumoniae and Haemophilus influenzae from healthy and sick individuals varies with age. J Clin Microbiol. 2004;42:4604–9. doi: 10.1128/JCM.42.10.4604-4609.2004. - DOI - PMC - PubMed
-
- Sulikowska A, Grzesiowski P, Sadowy E, Fiett J, Hryniewicz W. Characteristics of Streptococcus pneumoniae, Haemophilus influenzae, and Moraxella catarrhalis isolated from the nasopharynges of asymptomatic children and molecular analysis of S. pneumoniae and H. influenzae strain replacement in the nasopharynx. J Clin Microbiol. 2004;42:3942–9. doi: 10.1128/JCM.42.9.3942-3949.2004. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

