On consensus biomarker selection
- PMID: 17570864
- PMCID: PMC1892093
- DOI: 10.1186/1471-2105-8-S5-S5
On consensus biomarker selection
Abstract
Background: Recent development of mass spectrometry technology enabled the analysis of complex peptide mixtures. A lot of effort is currently devoted to the identification of biomarkers in human body fluids like serum or plasma, based on which new diagnostic tests for different diseases could be constructed. Various biomarker selection procedures have been exploited in recent studies. It has been noted that they often lead to different biomarker lists and as a consequence, the patient classification may also vary.
Results: Here we propose a new approach to the biomarker selection problem: to apply several competing feature ranking procedures and compute a consensus list of features based on their outcomes. We validate our methods on two proteomic datasets for the diagnosis of ovarian and prostate cancer.
Conclusion: The proposed methodology can improve the classification results and at the same time provide a unified biomarker list for further biological examinations and interpretation.
Figures

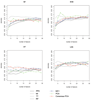


References
-
- Adam BL, Qu Y, Davis JW, Ward MD, Clements MA, Cazares LH, Semmes OJ, Schellhammer PF, Yasui Y, Feng Z, Wright GLJ. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Research. 2002;62:3609–3614. - PubMed
-
- Li J, Zhang Z, Rosenzweig J, Wang YY, Chan DW. Proteomics and bioinformatics approaches for identification of serum biomarkers to detect breast cancer. Clinical Chemistry. 2002;48:1296–1304. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

