Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus
- PMID: 17573971
- PMCID: PMC1925096
- DOI: 10.1186/1471-2164-8-174
Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus
Abstract
Background: The number of completely sequenced plastid genomes available is growing rapidly. This array of sequences presents new opportunities to perform comparative analyses. In comparative studies, it is often useful to compare across wide phylogenetic spans and, within angiosperms, to include representatives from basally diverging lineages such as the genomes reported here: Nuphar advena (from a basal-most lineage) and Ranunculus macranthus (a basal eudicot). We report these two new plastid genome sequences and make comparisons (within angiosperms, seed plants, or all photosynthetic lineages) to evaluate features such as the status of ycf15 and ycf68 as protein coding genes, the distribution of simple sequence repeats (SSRs) and longer dispersed repeats (SDR), and patterns of nucleotide composition.
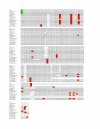
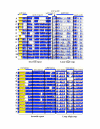
Results: The Nuphar [GenBank:NC_008788] and Ranunculus [GenBank:NC_008796] plastid genomes share characteristics of gene content and organization with many other chloroplast genomes. Like other plastid genomes, these genomes are A+T-rich, except for rRNA and tRNA genes. Detailed comparisons of Nuphar with Nymphaea, another Nymphaeaceae, show that more than two-thirds of these genomes exhibit at least 95% sequence identity and that most SSRs are shared. In broader comparisons, SSRs vary among genomes in terms of abundance and length and most contain repeat motifs based on A and T nucleotides.
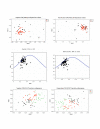
Conclusion: SSR and SDR abundance varies by genome and, for SSRs, is proportional to genome size. Long SDRs are rare in the genomes assessed. SSRs occur less frequently than predicted and, although the majority of the repeat motifs do include A and T nucleotides, the A+T bias in SSRs is less than that predicted from the underlying genomic nucleotide composition. In codon usage third positions show an A+T bias, however variation in codon usage does not correlate with differences in A+T-richness. Thus, although plastome nucleotide composition shows "A+T richness", an A+T bias is not apparent upon more in-depth analysis, at least in these aspects. The pattern of evolution in the sequences identified as ycf15 and ycf68 is not consistent with them being protein-coding genes. In fact, these regions show no evidence of sequence conservation beyond what is normal for non-coding regions of the IR.
Figures




References
-
- Chang C-C, Lin H-C, Lin I-P, Chow T-Y, Chen H-H, Chen W-H, Cheng C-H, Lin C-Y, Liu S-M, Chang C-C, Chaw S-M. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. - DOI - PubMed
Publication types
MeSH terms
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

