mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish
- PMID: 17574232
- PMCID: PMC2148033
- DOI: 10.1016/j.ydbio.2007.05.018
mef2ca is required in cranial neural crest to effect Endothelin1 signaling in zebrafish
Abstract
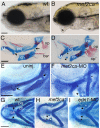
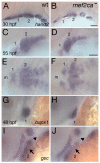
Mef2 genes encode highly conserved transcription factors involved in somitic and cardiac mesoderm development in diverse bilaterians. Vertebrates have multiple mef2 genes. In mice, mef2c is required for heart and vascular development. We show that a zebrafish mef2c gene (mef2ca) is required in cranial neural crest (CNC) for proper head skeletal patterning. mef2ca mutants have head skeletal phenotypes resembling those seen upon partial loss-of-function of endothelin1 (edn1). Furthermore, mef2ca interacts genetically with edn1, arguing that mef2ca functions within the edn1 pathway. mef2ca is expressed in CNC and this expression does not require edn1 signaling. Mosaic analyses reveal that mef2ca is required in CNC for pharyngeal skeletal morphogenesis. Proper expression of many edn1-dependent target genes including hand2, bapx1, and gsc, depends upon mef2ca function. mef2ca plays a critical role in establishing the proper nested expression patterns of dlx genes. dlx5a and dlx6a, known Edn1 targets, are downregulated in mef2ca mutant pharyngeal arch CNC. Surprisingly, dlx4b and dlx3b are oppositely affected in mef2ca mutants. dlx4b expression is abolished while the edn1-dependent dlx3b is ectopically expressed in more dorsal CNC. Together our results support a model in which CNC cells require mef2ca downstream of edn1 signaling for proper craniofacial development.
Figures







References
-
- Angelo S, et al. Conservation of sequence and expression of Xenopus and zebrafish dHAND during cardiac, branchial arch and lateral mesoderm development. Mech Dev. 2000;95:231–7. - PubMed
-
- Bi W, et al. The transcription factor MEF2C-null mouse exhibits complex vascular malformations and reduced cardiac expression of angiopoietin 1 and VEGF. Dev Biol. 1999;211:255–67. - PubMed
-
- Black BL, Olson EN. Transcriptional control of muscle development by myocyte enhancer factor-2 (MEF2) proteins. Annu Rev Cell Dev Biol. 1998;14:167–96. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

