Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis
- PMID: 17575002
- PMCID: PMC1951034
- DOI: 10.1128/AEM.00988-07
Transposon insertion reveals pRM, a plasmid of Rickettsia monacensis
Abstract
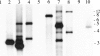
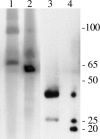
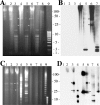
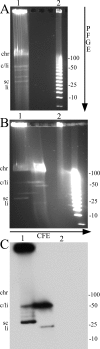
Until the recent discovery of pRF in Rickettsia felis, the obligate intracellular bacteria of the genus Rickettsia (Rickettsiales: Rickettsiaceae) were thought not to possess plasmids. We describe pRM, a plasmid from Rickettsia monacensis, which was detected by pulsed-field gel electrophoresis and Southern blot analyses of DNA from two independent R. monacensis populations transformed by transposon-mediated insertion of coupled green fluorescent protein and chloramphenicol acetyltransferase marker genes into pRM. Two-dimensional electrophoresis showed that pRM was present in rickettsial cells as circular and linear isomers. The 23,486-nucleotide (31.8% G/C) pRM plasmid was cloned from the transformant populations by chloramphenicol marker rescue of restriction enzyme-digested transformant DNA fragments and PCR using primers derived from sequences of overlapping restriction fragments. The plasmid was sequenced. Based on BLAST searches of the GenBank database, pRM contained 23 predicted genes or pseudogenes and was remarkably similar to the larger pRF plasmid. Two of the 23 genes were unique to pRM and pRF among sequenced rickettsial genomes, and 4 of the genes shared by pRM and pRF were otherwise found only on chromosomes of R. felis or the ancestral group rickettsiae R. bellii and R. canadensis. We obtained pulsed-field gel electrophoresis and Southern blot evidence for a plasmid in R. amblyommii isolate WB-8-2 that contained genes conserved between pRM and pRF. The pRM plasmid may provide a basis for the development of a rickettsial transformation vector.
Figures

References
-
- Akman, L., A. Yamashita, H. Watanabe, K. Oshima, T. Shiba, M. Hattori, and S. Aksoy. 2002. Genome sequence of the endocellular obligate symbiont of tsetse flies, Wigglesworthia glossinidia. Nat. Genet. 32:402-407. - PubMed
-
- Andersson, S. G., A. Zomorodipour, J. O. Andersson, T. Sicheritz-Ponten, U. C. Alsmark, R. M. Podowski, A. K. Naslund, A. Eriksson, H. H. Winkler, and C. G. Kurland. 1998. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396:133-140. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

