RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena
- PMID: 17575054
- PMCID: PMC1891430
- DOI: 10.1101/gad.1544207
RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena
Abstract
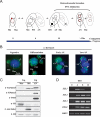
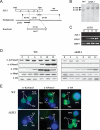
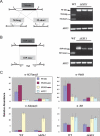
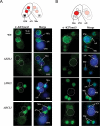
Methylated H3K27 is an important mark for Polycomb group (PcG) protein-mediated transcriptional gene silencing (TGS) in multicellular eukaryotes. Here a Drosophila E(z) homolog, EZL1, is characterized in the ciliated protozoan Tetrahymena thermophila and is shown to be responsible for H3K27 methylation associated with developmentally regulated heterochromatin formation and DNA elimination. Importantly, Ezl1p-catalyzed H3K27 methylation occurs in an RNA interference (RNAi)-dependent manner. H3K27 methylation also regulates H3K9 methylation in these processes. Furthermore, an "effector" of programmed DNA elimination, the chromodomain protein Pdd1p, is shown to bind both K27- and K9-methylated H3. These studies provide a framework for an RNAi-dependent, Polycomb group protein-mediated heterochromatin formation pathway in Tetrahymena and underscore the connection between the two highly conserved machineries in eukaryotes.
Figures
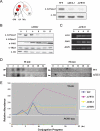

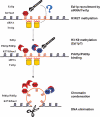
References
-
- Allis C.D., Bowen J.K., Abraham G.N., Glover C.V., Gorovsky M.A., Bowen J.K., Abraham G.N., Glover C.V., Gorovsky M.A., Abraham G.N., Glover C.V., Gorovsky M.A., Glover C.V., Gorovsky M.A., Gorovsky M.A. Proteolytic processing of histone H3 in chromatin: A physiologically regulated event in Tetrahymena micronuclei. Cell. 1980;20:55–64. - PubMed
-
- Baldauf S.L., Roger A.J., Wenk-Siefert I., Doolittle W.F., Roger A.J., Wenk-Siefert I., Doolittle W.F., Wenk-Siefert I., Doolittle W.F., Doolittle W.F. A kingdom-level phylogeny of eukaryotes based on combined protein data. Science. 2000;290:972–977. - PubMed
-
- Bannister A.J., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Zegerman P., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Partridge J.F., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Miska E.A., Thomas J.O., Allshire R.C., Kouzarides T., Thomas J.O., Allshire R.C., Kouzarides T., Allshire R.C., Kouzarides T., Kouzarides T. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. - PubMed
-
- Bastow R., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C., Mylne J.S., Lister C., Lippman Z., Martienssen R.A., Dean C., Lister C., Lippman Z., Martienssen R.A., Dean C., Lippman Z., Martienssen R.A., Dean C., Martienssen R.A., Dean C., Dean C. Vernalization requires epigenetic silencing of FLC by histone methylation. Nature. 2004;427:164–167. - PubMed
-
- Bender L.B., Cao R., Zhang Y., Strome S., Cao R., Zhang Y., Strome S., Zhang Y., Strome S., Strome S. The MES-2/MES-3/MES-6 complex and regulation of histone H3 methylation in C. elegans. Curr. Biol. 2004;14:1639–1643. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
