Intramolecular DNA quadruplexes with different arrangements of short and long loops
- PMID: 17576685
- PMCID: PMC1919480
- DOI: 10.1093/nar/gkm316
Intramolecular DNA quadruplexes with different arrangements of short and long loops
Abstract
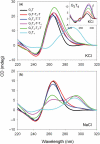
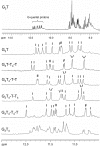
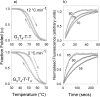
We have examined the folding, stability and kinetics of intramolecular quadruplexes formed by DNA sequences containing four G3 tracts separated by either single T or T4 loops. All these sequences fold to form intramolecular quadruplexes and 1D-NMR spectra suggest that they each adopt unique structures (with the exception of the sequence with all three loops containing T4, which is polymorphic). The stability increases with the number of single T loops, though the arrangement of different length loops has little effect. In the presence of potassium ions, the oligonucleotides that contain at least one single T loop exhibit similar CD spectra, which are indicative of a parallel topology. In contrast, when all three loops are substituted with T4 the CD spectrum is typical of an antiparallel arrangement. In the presence of sodium ions, the sequences with two and three single T loops also adopt a parallel folded structure. Kinetic studies on the complexes with one or two T4 loops in the presence of potassium ions reveal that sequences with longer loops display slower folding rates.
Figures

References
-
- Davis JT. G-quartets 40 years later: From 5′-GMP to molecular biology and supramolecular chemistry. Angew. Chem. Int. Ed. Engl. 2004;43:668–698. - PubMed
-
- Simonsson T. G-quadruplex DNA structures - Variations on a theme. Biol. Chem. 2001;382:621–628. - PubMed
-
- Sen D, Gilbert W. A sodium-potassium switch in the formation of 4-stranded G4-DNA. Nature. 1990;344:410–414. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

