Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR
- PMID: 17576689
- PMCID: PMC1935001
- DOI: 10.1093/nar/gkm443
Manipulation of P-TEFb control machinery by HIV: recruitment of P-TEFb from the large form by Tat and binding of HEXIM1 to TAR
Abstract
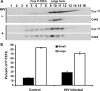
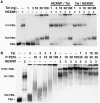
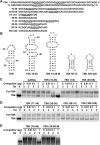
Basal transcription of the HIV LTR is highly repressed and requires Tat to recruit the positive transcription elongation factor, P-TEFb, which functions to promote the transition of RNA polymerase II from abortive to productive elongation. P-TEFb is found in two forms in cells, a free, active form and a large, inactive complex that also contains 7SK RNA and HEXIM1 or HEXIM2. Here we show that HIV infection of cells led to the release of P-TEFb from the large form. Consistent with Tat being the cause of this effect, transfection of a FLAG-tagged Tat in 293T cells caused a dramatic shift of P-TEFb out of the large form to a smaller form containing Tat. In vitro, Tat competed with HEXIM1 for binding to 7SK, blocked the formation of the P-TEFb-HEXIM1-7SK complex, and caused the release P-TEFb from a pre-formed P-TEFb-HEXIM1-7SK complex. These findings indicate that Tat can acquire P-TEFb from the large form. In addition, we found that HEXIM1 binds tightly to the HIV 5' UTR containing TAR and recruits and inhibits P-TEFb activity. This suggests that in the absence of Tat, HEXIM1 may bind to TAR and repress transcription elongation of the HIV LTR.
Figures




References
-
- Garriga J, Grana X. Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene. 2004;337:15–23. - PubMed
-
- Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol. Cell. 2006;23:297–305. - PubMed
-
- Shore SM, Byers SA, Maury W, Price DH. Identification of a novel isoform of Cdk9. Gene. 2003;307:175–182. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

