Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore
- PMID: 17576797
- PMCID: PMC2064361
- DOI: 10.1083/jcb.200702062
Spindly, a novel protein essential for silencing the spindle assembly checkpoint, recruits dynein to the kinetochore
Abstract
The eukaryotic spindle assembly checkpoint (SAC) monitors microtubule attachment to kinetochores and prevents anaphase onset until all kinetochores are aligned on the metaphase plate. In higher eukaryotes, cytoplasmic dynein is involved in silencing the SAC by removing the checkpoint proteins Mad2 and the Rod-Zw10-Zwilch complex (RZZ) from aligned kinetochores (Howell, B.J., B.F. McEwen, J.C. Canman, D.B. Hoffman, E.M. Farrar, C.L. Rieder, and E.D. Salmon. 2001. J. Cell Biol. 155:1159-1172; Wojcik, E., R. Basto, M. Serr, F. Scaerou, R. Karess, and T. Hays. 2001. Nat. Cell Biol. 3:1001-1007). Using a high throughput RNA interference screen in Drosophila melanogaster S2 cells, we have identified a new protein (Spindly) that accumulates on unattached kinetochores and is required for silencing the SAC. After the depletion of Spindly, dynein cannot target to kinetochores, and, as a result, cells arrest in metaphase with high levels of kinetochore-bound Mad2 and RZZ. We also identified a human homologue of Spindly that serves a similar function. However, dynein's nonkinetochore functions are unaffected by Spindly depletion. Our findings indicate that Spindly is a novel regulator of mitotic dynein, functioning specifically to target dynein to kinetochores.
Figures


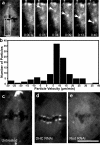


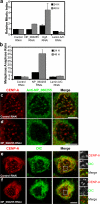
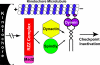
References
-
- Acquaviva, C., and J. Pines. 2006. The anaphase-promoting complex/cyclosome: APC/C. J. Cell Sci. 119:2401–2404. - PubMed
-
- Bailey, T.L., and C. Elkan. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2:28–36. - PubMed
-
- Bailey, T.L., and M. Gribskov. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics. 14:48–54. - PubMed
-
- Baker, D.J., J. Chen, and J.M. van Deursen. 2005. The mitotic checkpoint in cancer and aging: what have mice taught us? Curr. Opin. Cell Biol. 17:583–589. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

