Predictive genotypic algorithm for virologic response to lopinavir-ritonavir in protease inhibitor-experienced patients
- PMID: 17576846
- PMCID: PMC2043245
- DOI: 10.1128/AAC.00388-07
Predictive genotypic algorithm for virologic response to lopinavir-ritonavir in protease inhibitor-experienced patients
Erratum in
- Antimicrob Agents Chemother. 2008 Feb;52(2):811
Abstract
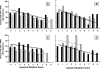
Several genotypic resistance algorithms have been proposed for quantitation of the degree of phenotypic resistance to the human immunodeficiency virus (HIV) protease inhibitor (PI) lopinavir (LPV), including the original LPV mutation score. In this study, we retrospectively evaluated 21 codons in HIV protease known to be associated with PI resistance in a large antiretroviral agent-experienced observational patient cohort, "Autorisation Temporaire d'Utilization" (ATU), to assess whether a more optimal algorithm could be derived by using virologic response data from patients treated with LPV in combination with ritonavir (LPV/r). Five of the 11 mutations constituting the LPV mutation score were not associated with a virologic response, while 4 additional mutations not included in this score demonstrated an association. Therefore, the LPV ATU score, which includes mutations at codons 10, 20, 24, 33, 36, 47, 48, 54, 82, and 84, was constructed and shown in two different types of multivariable analyses of the ATU cohort to be a better predictor of the virologic response than the LPV mutation score. The LPV ATU score was also more strongly associated with a virologic response when it was applied to independent clinical trial populations of PI-experienced patients receiving LPV/r. This study provides the basis for a new genotypic resistance algorithm that is useful for predicting the antiviral activities of LPV/r-based regimens in PI-experienced patients. The refined algorithm may be useful in making clinical treatment decisions and in refining genetic and pharmacologic methods for assessing the activity of LPV/r.
Figures


References
-
- Baxter, J. D., J. M. Schapiro, C. A. Boucher, V. M. Kohlbrenner, D. B. Hall, J. R. Scherer, and D. L. Mayers. 2006. Genotypic changes in human immunodeficiency virus type 1 protease associated with reduced susceptibility and virologic response to the protease inhibitor tipranavir. J. Virol. 80:10794-10801. - PMC - PubMed
-
- Benson, C. A., S. G. Deeks, S. C. Brun, R. M. Gulick, J. J. Eron, H. A. Kessler, R. L. Murphy, C. Hicks, M. King, D. Wheeler, J. Feinberg, R. Stryker, P. E. Sax, S. Riddler, M. Thompson, K. Real, A. Hsu, D. Kempf, A. J. Japour, and E. Sun. 2002. Safety and antiviral activity at 48 weeks of lopinavir/ritonavir plus nevirapine and 2 nucleoside reverse-transcriptase inhibitors in human immunodeficiency virus type 1-infected protease inhibitor-experienced patients. J. Infect. Dis. 185:599-607. - PubMed
-
- Brieman, L., J. Friedman, R. Olshen, and C. Stone. 1984. Classification and regression trees. Wadsworth International Group, Belmont, CA.
-
- Centers for Disease Control and Prevention. 1994. 1994 revised classification system for human immunodeficiency virus infection in children less than 13 years of age. Morb. Mortal. Wkly. Rep. 43(No. RR-12):1-10.
-
- de Mendoza, C., and V. Soriano. 2004. Resistance to HIV protease inhibitors: mechanisms and clinical consequences. Curr. Drug Metab. 5:321-328. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

