Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs
- PMID: 17577415
- PMCID: PMC1906771
- DOI: 10.1186/1471-2164-8-177
Towards the ictalurid catfish transcriptome: generation and analysis of 31,215 catfish ESTs
Abstract
Background: EST sequencing is one of the most efficient means for gene discovery and molecular marker development, and can be additionally utilized in both comparative genome analysis and evaluation of gene duplications. While much progress has been made in catfish genomics, large-scale EST resources have been lacking. The objectives of this project were to construct primary cDNA libraries, to conduct initial EST sequencing to generate catfish EST resources, and to obtain baseline information about highly expressed genes in various catfish organs to provide a guide for the production of normalized and subtracted cDNA libraries for large-scale transcriptome analysis in catfish.
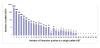
Results: A total of 17 cDNA libraries were constructed including 12 from channel catfish (Ictalurus punctatus) and 5 from blue catfish (I. furcatus). A total of 31,215 ESTs, with average length of 778 bp, were generated including 20,451 from the channel catfish and 10,764 from blue catfish. Cluster analysis indicated that 73% of channel catfish and 67% of blue catfish ESTs were unique within the project. Over 53% and 50% of the channel catfish and blue catfish ESTs, respectively, had significant similarities to known genes. All ESTs have been deposited in GenBank. Evaluation of the catfish EST resources demonstrated their potential for molecular marker development, comparative genome analysis, and evaluation of ancient and recent gene duplications. Subtraction of abundantly expressed genes in a variety of catfish tissues, identified here, will allow the production of low-redundancy libraries for in-depth sequencing.
Conclusion: The sequencing of 31,215 ESTs from channel catfish and blue catfish has significantly increased the EST resources in catfish. The EST resources should provide the potential for microarray development, polymorphic marker identification, mapping, and comparative genome analysis.
Figures

References
-
- USDA . Catfish Production Report. National Agricultural Statistics Service USDA Washington, D.C; (July 23, 2005)
-
- Chatakondi NG, Yant DR, Dunham RA. Commercial production and performance evaluation of channel catfish, Ictalurus punctatus female × blue catfish, Ictalurus furcatus male F-1 hybrids. Aquaculture. 2005;247:8.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

