Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1(10) hematopoietic stem cells
- PMID: 17577929
- PMCID: PMC2752385
- DOI: 10.1016/j.exphem.2007.02.013
Sustained in vitro trigger of self-renewal divisions in Hoxb4hiPbx1(10) hematopoietic stem cells
Abstract
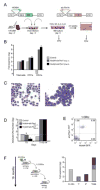
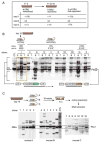
Factors that trigger and sustain self-renewal divisions in tissue stem cells remain poorly characterized. By modulating the levels of Hoxb4 and its co-factor Pbxl in primary hematopoietic cells (Hoxb4hiPbxl(10) cells), we report an in vitro expansion of mouse hematopoietic stem cells (HSCs) by 105-fold over 2 weeks, with subsequent preservation of HSC properties. Clonal analyses of the hematopoietic system in recipients of expanded HSCs indicate that up to 70% of Hoxb4hiPbxl(10) stem cells present at initiation of culture underwent self-renewal in vitro. In this setting, Hoxb4 and its co-factor did not promote an increase in DNA synthesis, or a decrease in doubling time of Scal+Lin- cells when compared to controls. Q-PCR analyses further revealed a downregulation of Cdknlb (p27Kipl) and Mxdl (MadI) transcript levels in Hoxb4hiPbxl(l0) primitive cells, accompanied by a more subtle increase in c-myc and reduction in Ccnd3 (Cyclin D3). We thus put forward this strategy as an efficient in vitro HSC expansion tool, enabling a further step into the avenue of self-renewal molecular effectors.
Figures




References
-
- Moore KA, Lemischka IR. Stem cells and their niches. Science. 2006;311:1880–1885. - PubMed
-
- Pawliuk R, Eaves C, Humphries RK. Evidence of both ontogeny and transplant dose-regulated expansion of hematopoietic stem cells in vivo. Blood. 1996;88:2852–2858. - PubMed
-
- Sauvageau G, Thorsteinsdottir U, Eaves CJ, et al. Overexpression of HOXB4 in hematopoietic cells causes the selective expansion of more primitive populations in vitro and in vivo. Genes Dev. 1995;9:1753–1765. - PubMed
-
- Iscove NN, Nawa K. Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol. 1997;7:805–808. - PubMed
-
- Thorsteinsdottir U, Sauvageau G, Humphries RK. Enhanced in vivo regenerative potential of HOXB4-transduced hematopoietic stem cells with regulation of their pool size. Blood. 1999;94:2605–2612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

