A multiresolution approach to automated classification of protein subcellular location images
- PMID: 17578580
- PMCID: PMC1933440
- DOI: 10.1186/1471-2105-8-210
A multiresolution approach to automated classification of protein subcellular location images
Abstract
Background: Fluorescence microscopy is widely used to determine the subcellular location of proteins. Efforts to determine location on a proteome-wide basis create a need for automated methods to analyze the resulting images. Over the past ten years, the feasibility of using machine learning methods to recognize all major subcellular location patterns has been convincingly demonstrated, using diverse feature sets and classifiers. On a well-studied data set of 2D HeLa single-cell images, the best performance to date, 91.5%, was obtained by including a set of multiresolution features. This demonstrates the value of multiresolution approaches to this important problem.
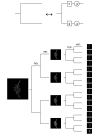
Results: We report here a novel approach for the classification of subcellular location patterns by classifying in multiresolution subspaces. Our system is able to work with any feature set and any classifier. It consists of multiresolution (MR) decomposition, followed by feature computation and classification in each MR subspace, yielding local decisions that are then combined into a global decision. With 26 texture features alone and a neural network classifier, we obtained an increase in accuracy on the 2D HeLa data set to 95.3%.
Conclusion: We demonstrate that the space-frequency localized information in the multiresolution subspaces adds significantly to the discriminative power of the system. Moreover, we show that a vastly reduced set of features is sufficient, consisting of our novel modified Haralick texture features. Our proposed system is general, allowing for any combinations of sets of features and any combination of classifiers.
Figures


References
-
- Boland M, Markey M, Murphy R. Proc IEEE Int Conf EMBS Society. Chicago, IL; 1997. Classification of Protein Localization Patterns Obtained via Fluorescence Light Microscopy; pp. 594–597.
-
- Boland M, Markey M, Murphy R. Automated Recognition of Patterns Characteristic of Subcellular Structures in Fluorescence Microscopy Images. Cytometry. 1998;33:366–375. - PubMed
-
- Boland M, Murphy R. A neural network classifier capable of recognizing the patterns of all major subcellular structures in fluorescence microscope images of HeLa cells. Bioinformatics. 2001;17:1213–1223. - PubMed
-
- Perner P, Perner H, Muller B. Mining Knowledge for Hep-2 Cell Image Classification. Journ Artificial Intelligence in Medicine. 2002;26:161–173. - PubMed
-
- Danckaert A, Gonzalez-Couto E, Bollondi L, Thompson N, Hayes B. Automated Recognition of Intracellular Organelles in Confocal Microscope Images. Traffic. 2002;3:66–73. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

