A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock
- PMID: 17578908
- PMCID: PMC1899476
- DOI: 10.1101/gad.1552207
A functional genomics strategy reveals clockwork orange as a transcriptional regulator in the Drosophila circadian clock
Abstract
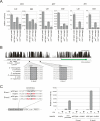
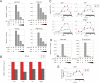
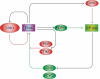
The Drosophila circadian clock consists of integrated autoregulatory feedback loops, making the clock difficult to elucidate without comprehensively identifying the network components in vivo. Previous studies have adopted genome-wide screening for clock-controlled genes using high-density oligonucleotide arrays that identified hundreds of clock-controlled genes. In an attempt to identify the core clock genes among these candidates, we applied genome-wide functional screening using an RNA interference (RNAi) system in vivo. Here we report the identification of novel clock gene candidates including clockwork orange (cwo), a transcriptional repressor belonging to the basic helix-loop-helix ORANGE family. cwo is rhythmically expressed and directly regulated by CLK-CYC through canonical E-box sequences. A genome-wide search for its target genes using the Drosophila genome tiling array revealed that cwo forms its own negative feedback loop and directly suppresses the expression of other clock genes through the E-box sequence. Furthermore, this negative transcriptional feedback loop contributes to sustaining a high-amplitude circadian oscillation in vivo. Based on these results, we propose that the competition between cyclic CLK-CYC activity and the adjustable threshold imposed by CWO keeps E-box-mediated transcription within the controllable range of its activity, thereby rendering a Drosophila circadian clock capable of generating high-amplitude oscillation.
Figures



References
-
- Allada R., White N.E., So W.V., Hall J.C., Rosbash M., White N.E., So W.V., Hall J.C., Rosbash M., So W.V., Hall J.C., Rosbash M., Hall J.C., Rosbash M., Rosbash M. A mutant Drosophila homolog of mammalian Clock disrupts circadian rhythms and transcription of period and timeless. Cell. 1998;93:791–804. - PubMed
-
- Blau J., Young M.W., Young M.W. Cycling vrille expression is required for a functional Drosophila clock. Cell. 1999;99:661–671. - PubMed
-
- Cawley S., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Bekiranov S., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Ng H.H., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kapranov P., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sekinger E.A., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Kampa D., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Piccolboni A., Sementchenko V., Cheng J., Williams A.J., Sementchenko V., Cheng J., Williams A.J., Cheng J., Williams A.J., Williams A.J., et al. Unbiased mapping of transcription factor binding sites along human chromosomes 21 and 22 points to widespread regulation of noncoding RNAs. Cell. 2004;116:499–509. - PubMed
-
- Ceriani M.F., Hogenesch J.B., Yanovsky M., Panda S., Straume M., Kay S.A., Hogenesch J.B., Yanovsky M., Panda S., Straume M., Kay S.A., Yanovsky M., Panda S., Straume M., Kay S.A., Panda S., Straume M., Kay S.A., Straume M., Kay S.A., Kay S.A. Genome-wide expression analysis in Drosophila reveals genes controlling circadian behavior. J. Neurosci. 2002;22:9305–9319. - PMC - PubMed
-
- Claridge-Chang A., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M.W., Wijnen H., Naef F., Boothroyd C., Rajewsky N., Young M.W., Naef F., Boothroyd C., Rajewsky N., Young M.W., Boothroyd C., Rajewsky N., Young M.W., Rajewsky N., Young M.W., Young M.W. Circadian regulation of gene expression systems in the Drosophila head. Neuron. 2001;32:657–671. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
