Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies
- PMID: 17578914
- PMCID: PMC1904155
- DOI: 10.1073/pnas.0704002104
Roles of DNA topoisomerase II isozymes in chemotherapy and secondary malignancies
Abstract
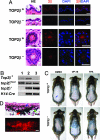
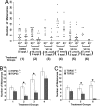
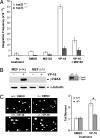
Drugs that target DNA topoisomerase II (Top2), including etoposide (VP-16), doxorubicin, and mitoxantrone, are among the most effective anticancer drugs in clinical use. However, Top2-based chemotherapy has been associated with higher incidences of secondary malignancies, notably the development of acute myeloid leukemia in VP-16-treated patients. This association is suggestive of a link between carcinogenesis and Top2-mediated DNA damage. We show here that VP-16-induced carcinogenesis involves mainly the beta rather than the alpha isozyme of Top2. In a mouse skin carcinogenesis model, the incidence of VP-16-induced melanomas in the skin of 7,12-dimethylbenz[a]anthracene-treated mice is found to be significantly higher in TOP2beta(+) than in skin-specific top2beta-knockout mice. Furthermore, VP-16-induced DNA sequence rearrangements and double-strand breaks (DSBs) are found to be Top2beta-dependent and preventable by cotreatment with a proteasome inhibitor, suggesting the importance of proteasomal degradation of the Top2beta-DNA cleavage complexes in VP-16-induced DNA sequence rearrangements. VP-16 cytotoxicity in transformed cells expressing both Top2 isozymes is, however, found to be primarily Top2alpha-dependent. These results point to the importance of developing Top2alpha-specific anticancer drugs for effective chemotherapy without the development of treatment-related secondary malignancies.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
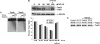

References
-
- Felix CA. Biochim Biophys Acta. 1998;1400:233–255. - PubMed
-
- Felix CA. Med Pediatr Oncol. 2001;36:525–535. - PubMed
-
- Pedersen-Bjergaard J, Andersen MK, Christiansen DH, Nerlov C. Blood. 2002;99:1909–1912. - PubMed
-
- Cimino G, Moir DT, Canaani O, Williams K, Crist WM, Katzav S, Cannizzaro L, Lange B, Nowell PC, Croce CM, et al. Cancer Res. 1991;51:6712–6714. - PubMed
-
- Djabali M, Selleri L, Parry P, Bower M, Young BD, Evans GA. Nat Genet. 1992;2:113–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

