Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit
- PMID: 17578920
- PMCID: PMC1904172
- DOI: 10.1073/pnas.0701967104
Monomeric G protein-coupled receptor rhodopsin in solution activates its G protein transducin at the diffusion limit
Abstract
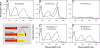
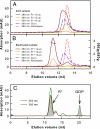
G protein-coupled receptors mediate biological signals by stimulating nucleotide exchange in heterotrimeric G proteins (Galphabetagamma). Receptor dimers have been proposed as the functional unit responsible for catalytic interaction with Galphabetagamma. To investigate whether a G protein-coupled receptor monomer can activate Galphabetagamma, we used the retinal photoreceptor rhodopsin and its cognate G protein transducin (G(t)) to determine the stoichiometry of rhodopsin/G(t) binding and the rate of catalyzed nucleotide exchange in G(t). Purified rhodopsin was prepared in dodecyl maltoside detergent solution. Rhodopsin was monomeric as concluded from fluorescence resonance energy transfer, copurification studies with fluorescent labeled and unlabeled rhodopsin, size exclusion chromatography, and multiangle laser light scattering. A 1:1 complex between light-activated rhodopsin and G(t) was found in the elution profiles, and one molecule of GDP was released upon complex formation. Analysis of the speed of catalytic rhodopsin/G(t) interaction yielded a maximum of approximately 50 G(t) molecules per second and molecule of activated rhodopsin. The bimolecular rate constant is close to the diffusion limit in the diluted system. The results show that the interaction of G(t) with an activated rhodopsin monomer is sufficient for fully functional G(t) activation. Although the activation rate in solution is at the physically possible limit, the rate in the native membrane is still 10-fold higher. This is likely attributable to the precise orientation of the G protein to the membrane surface, which enables a fast docking process preceding the actual activation step. Whether docking in membranes involves the formation of rhodopsin dimers or oligomers remains to be elucidated.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Foord SM, Bonner TI, Neubig RR, Rosser EM, Pin JP, Davenport AP, Spedding M, Harmar AJ. Pharmacol Rev. 2005;57:279–288. - PubMed
-
- Neves SR, Ram PT, Iyengar R. Science. 2002;296:1636–1639. - PubMed
-
- Liebman PA, Parker KR, Dratz EA. Annu Rev Physiol. 1987;49:765–791. - PubMed
-
- Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Nature. 2003;421:127–128. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

