Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65)
- PMID: 17579712
- PMCID: PMC1888729
- DOI: 10.1371/journal.pone.0000538
Control of pre-mRNA splicing by the general splicing factors PUF60 and U2AF(65)
Abstract
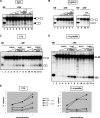
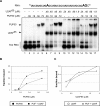
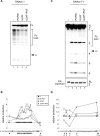
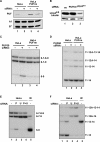
Pre-mRNA splicing is a crucial step in gene expression, and accurate recognition of splice sites is an essential part of this process. Splice sites with weak matches to the consensus sequences are common, though it is not clear how such sites are efficiently utilized. Using an in vitro splicing-complementation approach, we identified PUF60 as a factor that promotes splicing of an intron with a weak 3' splice-site. PUF60 has homology to U2AF(65), a general splicing factor that facilitates 3' splice-site recognition at the early stages of spliceosome assembly. We demonstrate that PUF60 can functionally substitute for U2AF(65)in vitro, but splicing is strongly stimulated by the presence of both proteins. Reduction of either PUF60 or U2AF(65) in cells alters the splicing pattern of endogenous transcripts, consistent with the idea that regulation of PUF60 and U2AF(65) levels can dictate alternative splicing patterns. Our results indicate that recognition of 3' splice sites involves different U2AF-like molecules, and that modulation of these general splicing factors can have profound effects on splicing.
Conflict of interest statement
Figures



References
-
- Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. - PubMed
-
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

