Relevance of the interaction between alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion
- PMID: 17581981
- PMCID: PMC1951408
- DOI: 10.1128/JVI.00900-07
Relevance of the interaction between alphaherpesvirus UL3.5 and UL48 proteins for virion maturation and neuroinvasion
Abstract
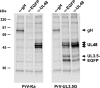
The UL3.5 and UL48 genes, which are conserved in most alphaherpesvirus genomes, are important for maturation of pseudorabies virus (PrV) particles in the cytoplasm of infected cells (W. Fuchs, B. G. Klupp, H. J. Rziha, and T. C. Mettenleiter, J. Virol. 70:3517-3527, 1996; W. Fuchs, H. Granzow, B. G. Klupp, M. Kopp and T. C. Mettenleiter, J. Virol. 76:6729-6742, 2002). In bovine herpesvirus 1 (BoHV-1), the homologous gene products pUL3.5 and pUL48 have been demonstrated to interact physically (N. Lam and G. Letchworth, J. Virol. 74:2876-2884, 2000). Moreover, BoHV-1 pUL3.5 partially complemented a pUL3.5 defect in PrV (W. Fuchs, H. Granzow, and T. C. Mettenleiter, J. Virol. 71:8886-8892, 1997). By using coimmunoprecipitation and yeast two-hybrid studies, we observed a similar interaction between pUL3.5 and pUL48 of PrV, as well as a heterologous interaction between the PrV and BoHV-1 gene products. The relevant domain could be confined to the first 43 amino acids of PrV pUL3.5. Unlike its BoHV-1 homologue, PrV pUL3.5 is processed by proteolytic cleavage, and only an abundant 14-kDa fragment consisting of amino acids 1 to >or=116 could be detected by peptide mass fingerprint analysis of purified wild-type PrV particles, which also contain the pUL48 tegument component. To determine the biological relevance of the protein-protein interaction, pUL3.5-, pUL48-, and double-negative PrV mutants were analyzed in parallel. All deletion mutants were replication competent but exhibited significantly reduced plaque sizes and virus titers in cultured rabbit kidney cells compared to wild-type and rescued viruses, which correlated with a delayed neuroinvasion in intranasally infected mice. Remarkably, the defects of the double-negative mutant were similar to those of pUL48-negative virus. Electron microscopy of cells infected with either deletion mutant revealed the retention of naked nucleocapsids in the cytoplasm and the absence of mature virus particles. In summary, our studies for the first time demonstrate the relevance of the pUL3.5-pUL48 interaction for secondary envelopment of an alphaherpesvirus, give a molecular basis for the observed trans-complementation between the PrV and BHV-1 pUL3.5 homologs, yield conclusive evidence for the incorporation of a proteolytically processed pUL3.5 into PrV virions, and demonstrate the importance of both proteins for neuroinvasion and neurovirulence of PrV.
Figures





References
-
- Campbell, M. E., J. W. Palfreyman, and C. M. Preston. 1984. Identification of herpes simplex virus DNA sequences which encode a trans-acting polypeptide responsible for stimulation of immediate early transcription. J. Mol. Biol. 180:1-19. - PubMed
-
- Cherepanov, P. P., and W. Wackernagel. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9-14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

