The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes
- PMID: 17584493
- PMCID: PMC2394765
- DOI: 10.1186/gb-2007-8-6-r119
The Sas3p and Gcn5p histone acetyltransferases are recruited to similar genes
Abstract
Background: Specific histone modifications can perform several cellular functions, for example, as signals to recruit trans-acting factors and as modulators of chromatin structure. Acetylation of Lys14 of histone H3 is the main target of many histone acetyltransferases in vitro and may play a central role in the stability of the nucleosome. This study is focused on the genome-wide binding of Saccharomyces cerevisiae histone acetyltransferases that are specific for Lys14 of histone H3.
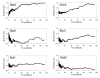
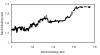
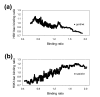
Results: We have used a variation of the genome-wide location analysis method, based on a macroarray platform, to identify binding sites of yeast histone acetyltransferase catalytic subunits and to correlate their positions with acetylation of Lys14 of histone H3. Our results revealed that the histone acetyltransferases Sas3p and Gcn5p are recruited to a pool of intensely transcribed genes and that there is considerable overlap between the two cohorts of Sas3p and Gcn5p bound gene pools. We also demonstrate a positive correlation between binding sites of both proteins and the acetylation state of Lys14 of histone H3. Finally, a positive correlation between the decrease of H3 Lys14 acetylation in a GCN5 deleted strain and the Gcn5p genome occupancy is shown.
Conclusion: Our data support a model in which both Gcn5p and Sas3p act as general activators of an overlapping pool of intensely transcribed genes. Since both proteins preferentially acetylate Lys14 of histone H3, our data support the hypothesis that acetylation of this specific residue facilitates the action of the transcriptional apparatus.
Figures


Similar articles
-
Histone H3 specific acetyltransferases are essential for cell cycle progression.Genes Dev. 2001 Dec 1;15(23):3144-54. doi: 10.1101/gad.931401. Genes Dev. 2001. PMID: 11731478 Free PMC article.
-
Acetylation of Rsc4p by Gcn5p is essential in the absence of histone H3 acetylation.Mol Cell Biol. 2008 Dec;28(23):6967-72. doi: 10.1128/MCB.00570-08. Epub 2008 Sep 22. Mol Cell Biol. 2008. PMID: 18809572 Free PMC article.
-
A conserved central region of yeast Ada2 regulates the histone acetyltransferase activity of Gcn5 and interacts with phospholipids.J Mol Biol. 2008 Dec 26;384(4):743-55. doi: 10.1016/j.jmb.2008.09.088. Epub 2008 Oct 11. J Mol Biol. 2008. PMID: 18950642
-
Histone acetylation, chromatin remodelling and nucleotide excision repair: hint from the study on MFA2 in Saccharomyces cerevisiae.Cell Cycle. 2005 Aug;4(8):1043-5. doi: 10.4161/cc.4.8.1928. Epub 2005 Aug 21. Cell Cycle. 2005. PMID: 16082210 Review.
-
Transcription: gene control by targeted histone acetylation.Curr Biol. 1998 Jun 4;8(12):R422-4. doi: 10.1016/s0960-9822(98)70268-4. Curr Biol. 1998. PMID: 9637914 Review.
Cited by
-
Balancing chromatin remodeling and histone modifications in transcription.Trends Genet. 2013 Nov;29(11):621-9. doi: 10.1016/j.tig.2013.06.006. Epub 2013 Jul 16. Trends Genet. 2013. PMID: 23870137 Free PMC article. Review.
-
The complex pattern of epigenomic variation between natural yeast strains at single-nucleosome resolution.Epigenetics Chromatin. 2015 Jul 31;8:26. doi: 10.1186/s13072-015-0019-3. eCollection 2015. Epigenetics Chromatin. 2015. PMID: 26229551 Free PMC article.
-
The prefoldin complex regulates chromatin dynamics during transcription elongation.PLoS Genet. 2013;9(9):e1003776. doi: 10.1371/journal.pgen.1003776. Epub 2013 Sep 19. PLoS Genet. 2013. PMID: 24068951 Free PMC article.
-
Promotion of Cell Viability and Histone Gene Expression by the Acetyltransferase Gcn5 and the Protein Phosphatase PP2A in Saccharomyces cerevisiae.Genetics. 2016 Aug;203(4):1693-707. doi: 10.1534/genetics.116.189506. Epub 2016 Jun 17. Genetics. 2016. PMID: 27317677 Free PMC article.
-
The Association of Rpb4 with RNA Polymerase II Depends on CTD Ser5P Phosphatase Rtr1 and Influences mRNA Decay in Saccharomyces cerevisiae.Int J Mol Sci. 2022 Feb 11;23(4):2002. doi: 10.3390/ijms23042002. Int J Mol Sci. 2022. PMID: 35216121 Free PMC article.
References
-
- Turner BM. Decoding the nucleosome. Cell. 1993;75:5–8. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

