The structure of the CstF-77 homodimer provides insights into CstF assembly
- PMID: 17584787
- PMCID: PMC1935011
- DOI: 10.1093/nar/gkm458
The structure of the CstF-77 homodimer provides insights into CstF assembly
Abstract
The cleavage stimulation factor (CstF) is essential for the first step of poly(A) tail formation at the 3' ends of mRNAs. This heterotrimeric complex is built around the 77-kDa protein bridging both CstF-64 and CstF-50 subunits. We have solved the crystal structure of the 77-kDa protein from Encephalitozoon cuniculi at a resolution of 2 A. The structure folds around 11 Half-a-TPR repeats defining two domains. The crystal structure reveals a tight homodimer exposing phylogenetically conserved areas for interaction with protein partners. Mapping experiments identify the C-terminal region of Rna14p, the yeast counterpart of CstF-77, as the docking domain for Rna15p, the yeast CstF-64 homologue.
Figures
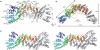
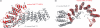

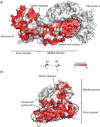

Similar articles
-
Crystal structure of murine CstF-77: dimeric association and implications for polyadenylation of mRNA precursors.Mol Cell. 2007 Mar 23;25(6):863-75. doi: 10.1016/j.molcel.2007.01.034. Mol Cell. 2007. PMID: 17386263
-
Locked tether formation by cooperative folding of Rna14p monkeytail and Rna15p hinge domains in the yeast CF IA complex.Structure. 2011 Apr 13;19(4):534-45. doi: 10.1016/j.str.2011.02.003. Structure. 2011. PMID: 21481776
-
Hexameric architecture of CstF supported by CstF-50 homodimerization domain structure.RNA. 2011 Mar;17(3):412-8. doi: 10.1261/rna.2481011. Epub 2011 Jan 13. RNA. 2011. PMID: 21233223 Free PMC article.
-
The structure of human cleavage factor I(m) hints at functions beyond UGUA-specific RNA binding: a role in alternative polyadenylation and a potential link to 5' capping and splicing.RNA Biol. 2011 Sep-Oct;8(5):748-53. doi: 10.4161/rna.8.5.16040. Epub 2011 Sep 1. RNA Biol. 2011. PMID: 21881408 Free PMC article. Review.
-
Structural biology of poly(A) site definition.Wiley Interdiscip Rev RNA. 2011 Sep-Oct;2(5):732-47. doi: 10.1002/wrna.88. Epub 2011 Apr 27. Wiley Interdiscip Rev RNA. 2011. PMID: 21823232 Free PMC article. Review.
Cited by
-
Integrative structural analysis of the UTPB complex, an early assembly factor for eukaryotic small ribosomal subunits.Nucleic Acids Res. 2016 Sep 6;44(15):7475-86. doi: 10.1093/nar/gkw562. Epub 2016 Jun 21. Nucleic Acids Res. 2016. PMID: 27330138 Free PMC article.
-
Reconstitution of CF IA from overexpressed subunits reveals stoichiometry and provides insights into molecular topology.Biochemistry. 2011 Nov 29;50(47):10203-14. doi: 10.1021/bi200964p. Epub 2011 Nov 2. Biochemistry. 2011. PMID: 22026644 Free PMC article.
-
Crystal structure of the Rna14-Rna15 complex.RNA. 2012 Jun;18(6):1154-62. doi: 10.1261/rna.032524.112. Epub 2012 Apr 18. RNA. 2012. PMID: 22513198 Free PMC article.
-
mRNA 3' end processing factors: a phylogenetic comparison.Comp Funct Genomics. 2012;2012:876893. doi: 10.1155/2012/876893. Epub 2012 Feb 6. Comp Funct Genomics. 2012. PMID: 22400011 Free PMC article.
-
Mutation of the polyadenylation complex subunit CstF77 reveals that mRNA 3' end formation and HSP101 levels are critical for a robust heat stress response.Plant Cell. 2023 Feb 20;35(2):924-941. doi: 10.1093/plcell/koac351. Plant Cell. 2023. PMID: 36472129 Free PMC article.
References
-
- Rosonina E, Kaneko S, Manley JL. Terminating the transcript: breaking up is hard to do. Genes Dev. 2006;20:1050–1056. - PubMed
-
- Proudfoot N. New perspectives on connecting messenger RNA 3′ end formation to transcription. Curr. Opin. Cell. Biol. 2004;16:272–278. - PubMed
-
- Edmonds M. A history of poly A sequences: from formation to factors to function. Prog. Nucleic Acid. Res. Mol. Biol. 2002;71:285–389. - PubMed
-
- Takagaki Y, Manley JL, MacDonald CC, Wilusz J, Shenk T. A multisubunit factor, CstF, is required for polyadenylation of mammalian pre-mRNAs. Genes Dev. 1990;4:2112–2120. - PubMed
-
- Minvielle-Sebastia L, Preker PJ, Keller W. RNA14 and RNA15 proteins as components of a yeast pre-mRNA 3′-end processing factor. Science. 1994;266:1702–1705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

