Sequence harmony: detecting functional specificity from alignments
- PMID: 17584793
- PMCID: PMC1933219
- DOI: 10.1093/nar/gkm406
Sequence harmony: detecting functional specificity from alignments
Abstract
Multiple sequence alignments are often used for the identification of key specificity-determining residues within protein families. We present a web server implementation of the Sequence Harmony (SH) method previously introduced. SH accurately detects subfamily specific positions from a multiple alignment by scoring compositional differences between subfamilies, without imposing conservation. The SH web server allows a quick selection of subtype specific sites from a multiple alignment given a subfamily grouping. In addition, it allows the predicted sites to be directly mapped onto a protein structure and displayed. We demonstrate the use of the SH server using the family of plant mitochondrial alternative oxidases (AOX). In addition, we illustrate the usefulness of combining sequence and structural information by showing that the predicted sites are clustered into a few distinct regions in an AOX homology model. The SH web server can be accessed at www.ibi.vu.nl/programs/seqharmwww.
Figures
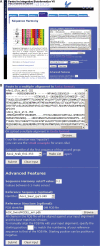


References
-
- Whisstock JC, Lesk AM. Prediction of protein function from protein sequence and structure. Quart. Rev. Biophys. 2003;36:307–340. - PubMed
-
- Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J. Mol. Biol. 2002;321:7–20. - PubMed
-
- Ye K, Lameijer E, Beukers MI, Jzerman A. A two-entropies analysis to identify functional positions in the transmembrane region of class A G protein-coupled receptors. Proteins. 2006;63:1018–1030. - PubMed

