Precise protein quantification based on peptide quantification using iTRAQ
- PMID: 17584939
- PMCID: PMC1940031
- DOI: 10.1186/1471-2105-8-214
Precise protein quantification based on peptide quantification using iTRAQ
Abstract
Background: Mass spectrometry based quantification of peptides can be performed using the iTRAQ reagent in conjunction with mass spectrometry. This technology yields information about the relative abundance of single peptides. A method for the calculation of reliable quantification information is required in order to obtain biologically relevant data at the protein expression level.
Results: A method comprising sound error estimation and statistical methods is presented that allows precise abundance analysis plus error calculation at the peptide as well as at the protein level. This yields the relevant information that is required for quantitative proteomics. Comparing the performance of our method named Quant with existing approaches the error estimation is reliable and offers information for precise bioinformatic models. Quant is shown to generate results that are consistent with those produced by ProQuant, thus validating both systems. Moreover, the results are consistent with that of Mascot 2.2. The MATLAB scripts of Quant are freely available via http://www.protein-ms.de and http://sourceforge.net/projects/protms/, each under the GNU Lesser General Public License.
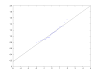
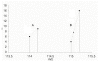
Conclusion: The software Quant demonstrates improvements in protein quantification using iTRAQ. Precise quantification data can be obtained at the protein level when using error propagation and adequate visualization. Quant integrates both and additionally provides the possibility to obtain more reliable results by calculation of wise quality measures. Peak area integration has been replaced by sum of intensities, yielding more reliable quantification results. Additionally, Quant allows the combination of quantitative information obtained by iTRAQ with peptide and protein identifications from popular tandem MS identification tools. Hence Quant is a useful tool for the proteomics community and may help improving analysis of proteomic experimental data. In addition, we have shown that a lognormal distribution fits the data of mass spectrometry based relative peptide quantification.
Figures






References
-
- NCBI National Center for Biotechnology Information http://www.ncbi.nih.gov/
-
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Mischoud K, O'Donovan C, Phan I, Pilbout S, Schneider M. The SWISS-PROT Protein Knowledgebase and Its Supplement TrEMBL in 2003. Nucleic Acids Research. 2003;31:365–370. doi: 10.1093/nar/gkg095. - DOI - PMC - PubMed
-
- Wilkins MR, Sanchez JC, Gooley AA, Appel RD, Humphery-Smith I, Hochstrasser DF, Williams KL. Progress with Proteome Projects: Why All Proteins Expressed by a Genome Should Be Identified and How to Do It. Biotechnology and Genetic Engineering Reviews. 1996. pp. 19–50. - PubMed
-
- Krijgsveld J, Heck AJR. Quantitative Proteomics by Metabolic Labelling with Stable Isotopes. Drug Discovery Today. 2004;3:S11–S15. doi: 10.1016/S1741-8372(04)02420-X. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

