BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth
- PMID: 17586627
- PMCID: PMC1951920
- DOI: 10.1128/JB.00331-07
BigR, a transcriptional repressor from plant-associated bacteria, regulates an operon implicated in biofilm growth
Abstract
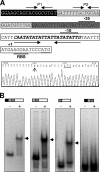
Xylella fastidiosa is a plant pathogen that colonizes the xylem vessels, causing vascular occlusion due to bacterial biofilm growth. However, little is known about the molecular mechanisms driving biofilm formation in Xylella-plant interactions. Here we show that BigR (for "biofilm growth-associated repressor") is a novel helix-turn-helix repressor that controls the transcription of an operon implicated in biofilm growth. This operon, which encodes BigR, membrane proteins, and an unusual beta-lactamase-like hydrolase (BLH), is restricted to a few plant-associated bacteria, and thus, we sought to understand its regulation and function in X. fastidiosa and Agrobacterium tumefaciens. BigR binds to a palindromic AT-rich element (the BigR box) in the Xylella and Agrobacterium blh promoters and strongly represses the transcription of the operon in these cells. The BigR box overlaps with two alternative -10 regions identified in the blh promoters, and mutations in this box significantly affected transcription, indicating that BigR competes with the RNA polymerase for the same promoter site. Although BigR is similar to members of the ArsR/SmtB family of regulators, our data suggest that, in contrast to the initial prediction, it does not act as a metal sensor. Increased activity of the BigR operon was observed in both Xylella and Agrobacterium biofilms. In addition, an A. tumefaciens bigR mutant showed constitutive expression of operon genes and increased biofilm formation on glass surfaces and tobacco roots, indicating that the operon may play a role in cell adherence or biofilm development.
Figures







References
-
- Brlansky, R. H., I. W. Timmer, W. J. French, and R. E. McCoy. 1983. Colonization of the sharpshooter vectors, Oncometopia nigricans and Homalodisca coagulata, by xylem-limited bacteria. Phytopathology 73:530-535.
-
- Busenlehner, L. S., M. A. Pennella, and D. P. Giedroc. 2003. The SmtB/ArsR family of metalloregulatory transcriptional repressors: structural insights into prokaryotic metal resistance. FEMS Microbiol. Rev. 27:131-143. - PubMed
-
- Chang, C. J., M. Garnier, L. Zreik, V. Rossetti, and J. M. Bové. 1993. Culture and serological detection of the xylem-limited bacterium causing citrus variegated chlorosis and its identification as a strain of Xylella fastidiosa. Curr. Microbiol. 27:137-142. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

