BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14
- PMID: 17586641
- PMCID: PMC2168662
- DOI: 10.1128/JB.00586-07
BifA, a cyclic-Di-GMP phosphodiesterase, inversely regulates biofilm formation and swarming motility by Pseudomonas aeruginosa PA14
Abstract
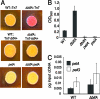
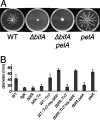
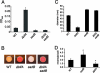
The intracellular signaling molecule, cyclic-di-GMP (c-di-GMP), has been shown to influence bacterial behaviors, including motility and biofilm formation. We report the identification and characterization of PA4367, a gene involved in regulating surface-associated behaviors in Pseudomonas aeruginosa. The PA4367 gene encodes a protein with an EAL domain, associated with c-di-GMP phosphodiesterase activity, as well as a GGDEF domain, which is associated with a c-di-GMP-synthesizing diguanylate cyclase activity. Deletion of the PA4367 gene results in a severe defect in swarming motility and a hyperbiofilm phenotype; thus, we designate this gene bifA, for biofilm formation. We show that BifA localizes to the inner membrane and, in biochemical studies, that purified BifA protein exhibits phosphodiesterase activity in vitro but no detectable diguanylate cyclase activity. Furthermore, mutational analyses of the conserved EAL and GGDEF residues of BifA suggest that both domains are important for the observed phosphodiesterase activity. Consistent with these data, the DeltabifA mutant exhibits increased cellular pools of c-di-GMP relative to the wild type and increased synthesis of a polysaccharide produced by the pel locus. This increased polysaccharide production is required for the enhanced biofilm formed by the DeltabifA mutant but does not contribute to the observed swarming defect. The DeltabifA mutation also results in decreased flagellar reversals. Based on epistasis studies with the previously described sadB gene, we propose that BifA functions upstream of SadB in the control of biofilm formation and swarming.
Figures

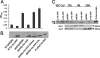


References
-
- Benz, R., and R. E. Hancock. 1981. Properties of the large ion-permeable pores formed from protein F of Pseudomonas aeruginosa in lipid bilayer membranes. Biochim. Biophys. Acta 646:298-308. - PubMed
-
- Bobrov, A. G., O. Kirillina, and R. D. Perry. 2005. The phosphodiesterase activity of the HmsP EAL domain is required for negative regulation of biofilm formation in Yersinia pestis. FEMS Microbiol. Lett. 247:123-130. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

