In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast
- PMID: 17586657
- PMCID: PMC1955713
- DOI: 10.1105/tpc.106.048744
In vivo visualization of Mg-protoporphyrin IX, a coordinator of photosynthetic gene expression in the nucleus and the chloroplast
Abstract
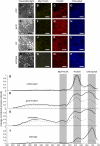
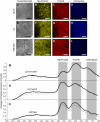
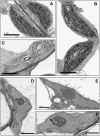
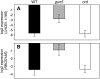
The photosynthetic apparatus is composed of proteins encoded by genes from both the nucleus and the chloroplast. To ensure that the photosynthetic complexes are assembled stoichiometrically and to enable their rapid reorganization in response to a changing environment, the plastids emit signals that regulate nuclear gene expression to match the status of the plastids. One of the plastid signals, the chlorophyll intermediate Mg-ProtoporphyrinIX (Mg-ProtoIX) accumulates under stress conditions and acts as a negative regulator of photosynthetic gene expression. By taking advantage of the photoreactive property of tetrapyrroles, Mg-ProtoIX could be visualized in the cells using confocal laser scanning spectroscopy. Our results demonstrate that Mg-ProtoIX accumulated both in the chloroplast and in the cytosol during stress conditions. Thus, the signaling metabolite is exported from the chloroplast, transmitting the plastid signal to the cytosol. Our results from the Mg-ProtoIX over- and underaccumulating mutants copper response defect and genome uncoupled5, respectively, demonstrate that the expression of both nuclear- and plastid-encoded photosynthesis genes is regulated by the accumulation of Mg-ProtoIX. Thus, stress-induced accumulation of the signaling metabolite Mg-ProtoIX coordinates nuclear and plastidic photosynthetic gene expression.
Figures






References
-
- Albrecht, V., Ingenfeld, A., and Apel, K. (2006). Characterization of the snowy cotyledon 1 mutant of Arabidopsis thaliana: The impact of chloroplast elongation factor G on chloroplast development and plant vitality. Plant Mol. Biol. 60 507–518. - PubMed
-
- Baba, K., Schmidt, J., Espinosa-Ruiz, A., Villarejo, A., Shiina, T., Gardestrom, P., Sane, A.P., and Bhalerao, R.P. (2004). Organellar gene transcription and early seedling development are affected in the rpoT;2 mutant of Arabidopsis. Plant J. 38 38–48. - PubMed
-
- Beck, C.F. (2005). Signaling pathways from the chloroplast to the nucleus. Planta 222 743–756. - PubMed
-
- Burgess, D., and Taylor, W. (1988). The chloroplast affects the transcription of a nuclear gene family. Mol. Gen. Genet. 214 89–96.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

