Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm
- PMID: 17586688
- PMCID: PMC1949899
- DOI: 10.1104/pp.107.102533
Characterization of SSIIIa-deficient mutants of rice: the function of SSIIIa and pleiotropic effects by SSIIIa deficiency in the rice endosperm
Abstract
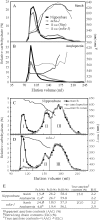
Starch synthase IIIa (SSIIIa)-deficient rice (Oryza sativa) mutants were generated using retrotransposon insertion and chemical mutagenesis. The lowest migrating SS activity bands on glycogen-containing native polyacrylamide gel, which were identified to be those for SSIIIa, were completely absent in these mutants, indicating that they are SSIIIa null mutants. The amylopectin B(2) to B(4) chains with degree of polymerization (DP) >/= 30 and the M(r) of amylopectin in the mutant were reduced to about 60% and 70% of the wild-type values, respectively, suggesting that SSIIIa plays an important part in the elongation of amylopectin B(2) to B(4) chains. Chains with DP 6 to 9 and DP 16 to 19 decreased while chains with DP 10 to 15 and DP 20 to 25 increased in the mutants amylopectin. These changes in the SSIIIa mutants are almost opposite images of those of SSI-deficient rice mutant and were caused by 1.3- to 1.7-fold increase of the amount of SSI in the mutants endosperm. Furthermore, the amylose content and the extralong chains (DP >/= 500) of amylopectin were increased by 1.3- and 12-fold, respectively. These changes in the composition in the mutants starch were caused by 1.4- to 1.7-fold increase in amounts of granules-bound starch synthase (GBSSI). The starch granules of the mutants were smaller with round shape, and were less crystalline. Thus, deficiency in SSIIIa, the second major SS isozyme in developing rice endosperm affected the structure of amylopectin, amylase content, and physicochemical properties of starch granules in two ways: directly by the SSIIIa deficiency itself and indirectly by the enhancement of both SSI and GBSSI gene transcripts.
Figures







References
-
- Abel GJW, Springer F, Willmitzer L, Kossman J (1996) Cloning and functional analysis of a cDNA encoding a novel 139 kDa starch synthase from potato (Solanum tuberosum L.). Plant J 10 981–991 - PubMed
-
- Ball SG, Morell MK (2003) From bacterial glycogen to starch: understanding the biogenesis of the plant starch granule. Annu Rev Plant Biol 54 207–233 - PubMed
-
- Boyer CD, Liu KC (1985) The interaction of endosperm genotype and genetic background. Starch/Stärke 37 73–79
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

