Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference
- PMID: 17586814
- PMCID: PMC1919482
- DOI: 10.1093/nar/gkm475
Conditional brain-specific knockdown of MAPK using Cre/loxP regulated RNA interference
Abstract
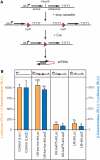
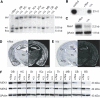
In the last years, RNA interference (RNAi)-mediated gene knockdown has developed into a routine method to assess gene function in cultured mammalian cells in a fast and easy manner. For the use of this technique in developing or adult mice, short hairpin (sh)RNA vectors expressed stably from the genome are a faster alternative to conventional knockout approaches. Here we describe an advanced strategy for conditional gene knockdown in mice, where we used the Cre/loxP system to activate RNAi in a time and tissue dependent manner in the adult mouse brain. By placing conditional RNAi constructs into the defined genomic Rosa26 locus and by using recombinase mediated cassette exchange (RMCE) instead of laborious homologous recombination, we developed a fast, easy and reproducible approach to assess gene function in adult mice. We applied this technique to three genes of the MAPK signaling pathway-Braf, Mek1 and Mek2-and demonstrate here the potential of this new tool in mouse mutagenesis.
Figures




References
-
- Dykxhoorn DM, Novina CD, Sharp PA. Killing the messenger: short RNAs that silence gene expression. Nat. Rev. Mol. Cell Biol. 2003;4:457–467. - PubMed
-
- Kunath T, Gish G, Lickert H, Jones N, Pawson T, Rossant J. Transgenic RNA interference in ES cell-derived embryos recapitulates a genetic null phenotype. Nat. Biotechnol. 2003;21:559–561. - PubMed
-
- Chen Y, Stamatoyannopoulos G, Song CZ. Down-regulation of CXCR4 by inducible small interfering RNA inhibits breast cancer cell invasion in vitro. Cancer Res. 2003;63:4801–4804. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

