Novel activity of eukaryotic translocase, eEF2: dissociation of the 80S ribosome into subunits with ATP but not with GTP
- PMID: 17586816
- PMCID: PMC1950535
- DOI: 10.1093/nar/gkm468
Novel activity of eukaryotic translocase, eEF2: dissociation of the 80S ribosome into subunits with ATP but not with GTP
Erratum in
- Nucleic Acids Res. 2007;35(20):7040
Abstract
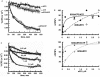
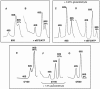
Ribosomes must dissociate into subunits in order to begin protein biosynthesis. The enzymes that catalyze this fundamental process in eukaryotes remained unknown. Here, we demonstrate that eukaryotic translocase, eEF2, which catalyzes peptide elongation in the presence of GTP, dissociates yeast 80S ribosomes into subunits in the presence of ATP but not GTP or other nucleoside triphosphates. Dissociation was detected by light scattering or ultracentrifugation after the split subunits were stabilized. ATP was hydrolyzed during the eEF2-dependent dissociation, while a non-hydrolyzable analog of ATP was inactive in ribosome splitting by eEF2. GTP inhibited not only ATP hydrolysis but also dissociation. Sordarin, a fungal eEF2 inhibitor, averted the splitting but stimulated ATP hydrolysis. Another elongation inhibitor, cycloheximide, also prevented eEF2/ATP-dependent splitting, while the inhibitory effect of fusidic acid on the splitting was nominal. Upon dissociation of the 80S ribosome, eEF2 was found on the subunits. We propose that the dissociation activity of eEF2/ATP plays a role in mobilizing 80S ribosomes for protein synthesis during the shift up of physiological conditions.
Figures


References
-
- Uesono Y, Toh EA. Transient inhibition of translation initiation by osmotic stress. J. Biol. Chem. 2002;277:13848–13855. - PubMed
-
- Volarevic S, Stewart MJ, Ledermann B, Zilberman F, Terracciano L, Montini E, Grompe M, Kozma SC, Thomas G. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

