Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome
- PMID: 17588519
- PMCID: PMC2031223
- DOI: 10.1016/j.molcel.2007.05.022
Ubiquitin-independent degradation of cell-cycle inhibitors by the REGgamma proteasome
Abstract
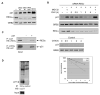
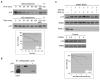
The cell-cycle regulator p21(Cip1) is degraded by proteasomes independently of ubiquitination. We now show that degradation of p21 in vivo does not require the 19S proteasome lid, which contains the ubiquitin-binding subunit. Instead, the major proteasomal pathway for p21 degradation involves an alternative proteasome lid, the REGgamma complex. REGgamma binds to p21 in vivo, and deletion of p21's REGgamma-binding site greatly extends its half-life. Knockdown of REGgamma by RNA interference stabilizes p21, p21 has a significantly extended half-life in REGgamma(-/-) murine embryonic fibroblasts, and the p21 abundance is elevated in REGgamma(-/-) mice. The role of REGgamma in cell-cycle regulation may extend beyond p21 regulation, because p16(INK4A) and p19(Arf) also bind to REGgamma and are stabilized in REGgamma-deficient cells.
Figures




References
-
- Barton LF, Runnels HA, Schell TD, Cho Y, Gibbons R, Tevethia SS, Deepe GS, Jr, Monaco JJ. Immune defects in 28-kDa proteasome activator gamma-deficient mice. J Immunol. 2004;172:3948–3954. - PubMed
-
- Ben-Saadon R, Fajerman I, Ziv T, Hellman U, Schwartz AL, Ciechanover A. The tumor suppressor protein p16 (INK4a) and the human papillomavirus oncoprotein-58 E7 are naturally occurring lysine-less proteins that are degraded by the ubiquitin system. Direct evidence for ubiquitination at the N-terminal residue. J Biol Chem. 2004;279:41414–41421. - PubMed
-
- Bendjennat M, Boulaire J, Jascur T, Brickner H, Barbier V, Sarasin A, Fotedar A, Fotedar R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. - PubMed
-
- Bloom J, Amador V, Bartolini F, DeMartino G, Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitination. Cell. 2003;115:71–82. - PubMed
-
- Chen X, Chi Y, Bloecher A, Aebersold R, Clurman BE, Roberts JM. N-acetylation and ubiquitin-independent proteasomal degradation of p21(Cip1) Mol Cell. 2004;16:839–847. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

