trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing
- PMID: 17588521
- PMCID: PMC1973159
- DOI: 10.1016/j.molcel.2007.05.020
trans-splicing to spliceosomal U2 snRNA suggests disruption of branch site-U2 pairing during pre-mRNA splicing
Abstract
Pairing between U2 snRNA and the branch site of spliceosomal introns is essential for spliceosome assembly and is thought to be required for the first catalytic step of splicing. We have identified an RNA comprising the 5' end of U2 snRNA and the 3' exon of the ACT1-CUP1 reporter gene, resulting from a trans-splicing reaction in which a 5' splice site-like sequence in the universally conserved branch site-binding region of U2 is used in trans as a 5' splice site for both steps of splicing in vivo. Formation of this product occurs in functional spliceosomes assembled on reporter genes whose 5' splice sites are predicted to bind poorly at the spliceosome catalytic center. Multiple spatially disparate splice sites in U2 can be used, calling into question both the fate of its pairing to the branch site and the details of its role in splicing catalysis.
Figures
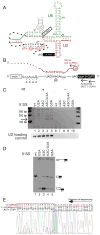



References
-
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosomes. In: Gesteland RF, Cech TR, Atkins JF, editors. The RNA World. 2. New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560.
-
- Konarska MM, Vilardell J, Query CC. Repositioning of the reaction intermediate within the catalytic center of the spliceosome. Mol Cell. 2006;21:543–553. - PubMed
-
- Madhani HD, Guthrie C. A novel base-pairing interaction between U2 and U6 snRNAs suggests a mechanism for the catalytic activation of the spliceosome. Cell. 1992;71:803–817. - PubMed
-
- Makarov EM, Makarova OV, Urlaub H, Gentzel M, Will CL, Wilm M, Lührmann R. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science. 2002;298:2205–2208. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

