Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis
- PMID: 17590082
- PMCID: PMC1894825
- DOI: 10.1371/journal.ppat.0030087
Transcriptional regulation of multi-drug tolerance and antibiotic-induced responses by the histone-like protein Lsr2 in M. tuberculosis
Abstract
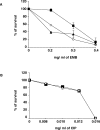
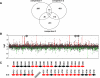
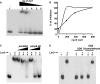
Multi-drug tolerance is a key phenotypic property that complicates the sterilization of mammals infected with Mycobacterium tuberculosis. Previous studies have established that iniBAC, an operon that confers multi-drug tolerance to M. bovis BCG through an associated pump-like activity, is induced by the antibiotics isoniazid (INH) and ethambutol (EMB). An improved understanding of the functional role of antibiotic-induced genes and the regulation of drug tolerance may be gained by studying the factors that regulate antibiotic-mediated gene expression. An M. smegmatis strain containing a lacZ gene fused to the promoter of M. tuberculosis iniBAC (PiniBAC) was subjected to transposon mutagenesis. Mutants with constitutive expression and increased EMB-mediated induction of PiniBAC::lacZ mapped to the lsr2 gene (MSMEG6065), a small basic protein of unknown function that is highly conserved among mycobacteria. These mutants had a marked change in colony morphology and generated a new polar lipid. Complementation with multi-copy M. tuberculosis lsr2 (Rv3597c) returned PiniBAC expression to baseline, reversed the observed morphological and lipid changes, and repressed PiniBAC induction by EMB to below that of the control M. smegmatis strain. Microarray analysis of an lsr2 knockout confirmed upregulation of M. smegmatis iniA and demonstrated upregulation of genes involved in cell wall and metabolic functions. Fully 121 of 584 genes induced by EMB treatment in wild-type M. smegmatis were upregulated ("hyperinduced") to even higher levels by EMB in the M. smegmatis lsr2 knockout. The most highly upregulated genes and gene clusters had adenine-thymine (AT)-rich 5-prime untranslated regions. In M. tuberculosis, overexpression of lsr2 repressed INH-mediated induction of all three iniBAC genes, as well as another annotated pump, efpA. The low molecular weight and basic properties of Lsr2 (pI 10.69) suggested that it was a histone-like protein, although it did not exhibit sequence homology with other proteins in this class. Consistent with other histone-like proteins, Lsr2 bound DNA with a preference for circular DNA, forming large oligomers, inhibited DNase I activity, and introduced a modest degree of supercoiling into relaxed plasmids. Lsr2 also inhibited in vitro transcription and topoisomerase I activity. Lsr2 represents a novel class of histone-like proteins that inhibit a wide variety of DNA-interacting enzymes. Lsr2 appears to regulate several important pathways in mycobacteria by preferentially binding to AT-rich sequences, including genes induced by antibiotics and those associated with inducible multi-drug tolerance. An improved understanding of the role of lsr2 may provide important insights into the mechanisms of action of antibiotics and the way that mycobacteria adapt to stresses such as antibiotic treatment.
Conflict of interest statement
Figures

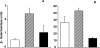






References
-
- Alland D, Kramnik I, Weisbrod TR, Otsubo L, Cerny R, et al. Identification of differentially expressed mRNA in prokaryotic organisms by customized amplification libraries (DECAL): The effect of isoniazid on gene expression in Mycobacterium tuberculosis . Proc Natl Acad Sci U S A. 1998;95:13227–13232. - PMC - PubMed
-
- Colangeli R, Helb D, Sridharan S, Sun J, Varma-Basil M, et al. The Mycobacterium tuberculosis iniA gene is essential for activity of an efflux pump that confers drug tolerance to both isoniazid and ethambutol. Mol Microbiol. 2005;55:1829–1840. - PubMed
-
- Balaban NQ, Merrin J, Chait R, Kowalik L, Leibler S. Bacterial persistence as a phenotypic switch. Science. 2004;305:1622–1625. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

