Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans
- PMID: 17590087
- PMCID: PMC1892049
- DOI: 10.1371/journal.pgen.0030108
Deletion at ITPR1 underlies ataxia in mice and spinocerebellar ataxia 15 in humans
Abstract
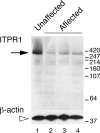
We observed a severe autosomal recessive movement disorder in mice used within our laboratory. We pursued a series of experiments to define the genetic lesion underlying this disorder and to identify a cognate disease in humans with mutation at the same locus. Through linkage and sequence analysis we show here that this disorder is caused by a homozygous in-frame 18-bp deletion in Itpr1 (Itpr1(Delta18/Delta18)), encoding inositol 1,4,5-triphosphate receptor 1. A previously reported spontaneous Itpr1 mutation in mice causes a phenotype identical to that observed here. In both models in-frame deletion within Itpr1 leads to a decrease in the normally high level of Itpr1 expression in cerebellar Purkinje cells. Spinocerebellar ataxia 15 (SCA15), a human autosomal dominant disorder, maps to the genomic region containing ITPR1; however, to date no causal mutations had been identified. Because ataxia is a prominent feature in Itpr1 mutant mice, we performed a series of experiments to test the hypothesis that mutation at ITPR1 may be the cause of SCA15. We show here that heterozygous deletion of the 5' part of the ITPR1 gene, encompassing exons 1-10, 1-40, and 1-44 in three studied families, underlies SCA15 in humans.
Conflict of interest statement
Competing interests. The authors have declared that no competing interests exist.
Figures



References
-
- Knight MA, Kennerson ML, Anney RJ, Matsuura T, Nicholson GA, et al. Spinocerebellar ataxia type 15 (SCA15) maps to 3p24.2–3pter: Exclusion of the ITPR1 gene, the human orthologue of an ataxic mouse mutant. Neurobiol Dis. 2003;13:147–157. - PubMed
-
- Davisson MT. Discovery genetics: Serendipity in basic research. ILAR J. 2005;46:338–345. - PubMed
-
- Matsumoto M, Nakagawa T, Inoue T, Nagata E, Tanaka K, et al. Ataxia and epileptic seizures in mice lacking type 1 inositol 1,4,5-trisphosphate receptor. Nature. 1996;379:168–171. - PubMed
-
- Storey E, Gardner RJM, Knight MA, Kennerson ML, Tuck RR, et al. A new autosomal dominant pure cerebellar ataxia. Neurology. 2001;57:1913–1915. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

