A role for Rab5 in structuring the endoplasmic reticulum
- PMID: 17591921
- PMCID: PMC2064419
- DOI: 10.1083/jcb.200701139
A role for Rab5 in structuring the endoplasmic reticulum
Erratum in
- J Cell Biol. 2007 Sep 24;178(7):1309
Abstract
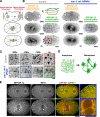
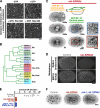
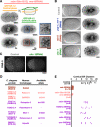
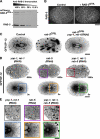
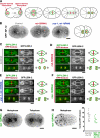
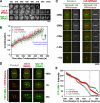
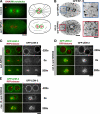
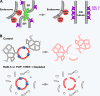
The endoplasmic reticulum (ER) is a contiguous network of interconnected membrane sheets and tubules. The ER is differentiated into distinct domains, including the peripheral ER and nuclear envelope. Inhibition of two ER proteins, Rtn4a and DP1/NogoA, was previously shown to inhibit the formation of ER tubules in vitro. We show that the formation of ER tubules in vitro also requires a Rab family GTPase. Characterization of the 29 Caenorhabditis elegans Rab GTPases reveals that depletion of RAB-5 phenocopies the defects in peripheral ER structure that result from depletion of RET-1 and YOP-1, the C. elegans homologues of Rtn4a and DP1/NogoA. Perturbation of endocytosis by other means did not affect ER structure; the role of RAB-5 in ER morphology is thus independent of its well-studied requirement for endocytosis. RAB-5 and YOP-1/RET-1 also control the kinetics of nuclear envelope disassembly, which suggests an important role for the morphology of the peripheral ER in this process.
Figures
Similar articles
-
Clathrin interaction and subcellular localization of Ce-DAB-1, an adaptor for protein secretion in Caenorhabditis elegans.Traffic. 2006 Mar;7(3):324-36. doi: 10.1111/j.1600-0854.2006.00386.x. Traffic. 2006. PMID: 16497226
-
Dynamics and inheritance of the endoplasmic reticulum.J Cell Sci. 2004 Jun 15;117(Pt 14):2871-8. doi: 10.1242/jcs.01286. J Cell Sci. 2004. PMID: 15197242 Review.
-
How the ER stays in shape.Cell. 2006 Feb 10;124(3):464-6. doi: 10.1016/j.cell.2006.01.017. Cell. 2006. PMID: 16469692
-
The maintenance of the endoplasmic reticulum network is regulated by p47, a cofactor of p97, through phosphorylation by cdc2 kinase.Genes Cells. 2005 Apr;10(4):333-44. doi: 10.1111/j.1365-2443.2005.00837.x. Genes Cells. 2005. PMID: 15773896
-
Molecular basis for sculpting the endoplasmic reticulum membrane.Int J Biochem Cell Biol. 2012 Sep;44(9):1436-43. doi: 10.1016/j.biocel.2012.05.013. Epub 2012 May 26. Int J Biochem Cell Biol. 2012. PMID: 22640864 Review.
Cited by
-
An ABCG Transporter Functions in Rab Localization and Lysosome-Related Organelle Biogenesis in Caenorhabditis elegans.Genetics. 2020 Feb;214(2):419-445. doi: 10.1534/genetics.119.302900. Epub 2019 Dec 17. Genetics. 2020. PMID: 31848222 Free PMC article.
-
Rab32 modulates apoptosis onset and mitochondria-associated membrane (MAM) properties.J Biol Chem. 2010 Oct 8;285(41):31590-602. doi: 10.1074/jbc.M110.101584. Epub 2010 Jul 29. J Biol Chem. 2010. PMID: 20670942 Free PMC article.
-
The nuclear envelope.Cold Spring Harb Perspect Biol. 2010 Mar;2(3):a000539. doi: 10.1101/cshperspect.a000539. Cold Spring Harb Perspect Biol. 2010. PMID: 20300205 Free PMC article. Review.
-
Crosstalk between the Rho and Rab family of small GTPases in neurodegenerative disorders.Front Cell Neurosci. 2023 Jan 27;17:1084769. doi: 10.3389/fncel.2023.1084769. eCollection 2023. Front Cell Neurosci. 2023. PMID: 36779014 Free PMC article. Review.
-
EXOSC10 sculpts the transcriptome during the growth-to-maturation transition in mouse oocytes.Nucleic Acids Res. 2020 Jun 4;48(10):5349-5365. doi: 10.1093/nar/gkaa249. Nucleic Acids Res. 2020. PMID: 32313933 Free PMC article.
References
-
- Baumann, O., and B. Walz. 2001. Endoplasmic reticulum of animal cells and its organization into structural and functional domains. Int. Rev. Cytol. 205:149–214. - PubMed
-
- Bobinnec, Y., C. Marcaillou, X. Morin, and A. Debec. 2003. Dynamics of the endoplasmic reticulum during early development of Drosophila melanogaster. Cell Motil. Cytoskeleton. 54:217–225. - PubMed
-
- Calero, M., G.R. Whittaker, and R.N. Collins. 2001. Yop1p, the yeast homolog of the polyposis locus protein 1, interacts with Yip1p and negatively regulates cell growth. J. Biol. Chem. 276:12100–12112. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

