Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis
- PMID: 17591937
- PMCID: PMC1899414
- DOI: 10.2353/jmoldx.2007.060167
Optimization of quantitative MGMT promoter methylation analysis using pyrosequencing and combined bisulfite restriction analysis
Abstract
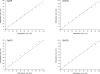
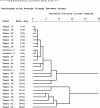
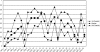
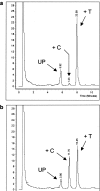
Resistance to chemotherapy is a major complication during treatment of cancer patients. Hypermethylation of the MGMT gene alters DNA repair and is associated with longer survival of glioblastoma patients treated with alkylating agents. Therefore, MGMT promoter methylation plays an important role as a predictive biomarker for chemotherapy resistance. To adopt this established correlation into a molecular diagnosis procedure, we compared and optimized three experimental techniques [combined bisulfite restriction analysis, a primer extension- and denaturing high-performance liquid chromatography-based method named SIRPH (SNuPE ion pair-reverse phase high-performance liquid chromatography), and pyrosequencing] with regard to their accuracy of detecting MGMT promoter methylation. Initially, bisulfite sequencing was used to obtain a comprehensive methylation profile of the MGMT promoter region in 22 glioblastoma samples and in three normal brain controls. Next, we statistically identified CpG sites that best discriminate between methylated and unmethylated MGMT promoters. These results were then used to design optimal combined bisulfite restriction analysis, SIRPH, and pyrosequencing assays for accurate and cost-efficient assessment of MGMT promoter methylation. We compared all three techniques with regard to their reliability and reproducibility on well-characterized tumor samples. The optimized pyrosequencing assay performed best and provides a sensitive, robust, and easy-to-use method for quantitative assessment of MGMT methylation, for both snap-frozen and paraffin-embedded specimens.
Figures





References
-
- Ludlum DB. DNA alkylation by the haloethylnitrosoureas: nature of modifications produced and their enzymatic repair or removal. Mutat Res. 1990;233:117–126. - PubMed
-
- Pegg AE. Repair of O6-alkylguanine by alkyltransferases. Mutat Res. 2000;462:83–100. - PubMed
-
- Pegg AE, Dolan ME, Moschel RC. Structure, function, and inhibition of O6-alkylguanine-DNA alkyltransferase. Prog Nucleic Acid Res Mol Biol. 1995;51:167–223. - PubMed
-
- Karran P, Bignami M. DNA damage tolerance, mismatch repair and genome instability. Bioessays. 1994;16:833–839. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

