Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation
- PMID: 17592110
- PMCID: PMC2040888
- DOI: 10.1073/pnas.0704769104
Structure of pyrrolysyl-tRNA synthetase, an archaeal enzyme for genetic code innovation
Abstract
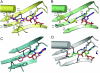
Pyrrolysine (Pyl), the 22nd natural amino acid and genetically encoded by UAG, becomes attached to its cognate tRNA by pyrrolysyl-tRNA synthetase (PylRS). We have determined three crystal structures of the Methanosarcina mazei PylRS complexed with either AMP-PNP, Pyl-AMP plus pyrophosphate, or the Pyl analogue N-epsilon-[(cylopentyloxy)carbonyl]-L-lysine plus ATP. The structures reveal that PylRS utilizes a deep hydrophobic pocket for recognition of the Pyl side chain. A comparison of these structures with previously determined class II tRNA synthetase complexes illustrates that different substrate specificities derive from changes in a small number of residues that form the substrate side-chain-binding pocket. The knowledge of these structures allowed the placement of PylRS in the aminoacyl-tRNA synthetase (aaRS) tree as the last known synthetase that evolved for genetic code expansion, as well as the finding that Pyl arose before the last universal common ancestral state. The PylRS structure provides an excellent framework for designing new aaRSs with altered amino acid specificity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
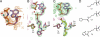

Similar articles
-
Recognition of non-alpha-amino substrates by pyrrolysyl-tRNA synthetase.J Mol Biol. 2009 Feb 6;385(5):1352-60. doi: 10.1016/j.jmb.2008.11.059. Epub 2008 Dec 11. J Mol Biol. 2009. PMID: 19100747
-
Crystallographic studies on multiple conformational states of active-site loops in pyrrolysyl-tRNA synthetase.J Mol Biol. 2008 May 2;378(3):634-52. doi: 10.1016/j.jmb.2008.02.045. Epub 2008 Feb 29. J Mol Biol. 2008. PMID: 18387634
-
Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.Cell Chem Biol. 2019 Jul 18;26(7):936-949.e13. doi: 10.1016/j.chembiol.2019.03.008. Epub 2019 Apr 25. Cell Chem Biol. 2019. PMID: 31031143
-
Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool.Biochim Biophys Acta. 2014 Jun;1844(6):1059-70. doi: 10.1016/j.bbapap.2014.03.002. Epub 2014 Mar 12. Biochim Biophys Acta. 2014. PMID: 24631543 Free PMC article. Review.
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
Cited by
-
Genetic Incorporation of ε-N-2-Hydroxyisobutyryl-lysine into Recombinant Histones.ACS Chem Biol. 2015 Jul 17;10(7):1599-603. doi: 10.1021/cb501055h. Epub 2015 Apr 29. ACS Chem Biol. 2015. PMID: 25909834 Free PMC article.
-
Enzymic recognition of amino acids drove the evolution of primordial genetic codes.Nucleic Acids Res. 2024 Jan 25;52(2):558-571. doi: 10.1093/nar/gkad1160. Nucleic Acids Res. 2024. PMID: 38048305 Free PMC article.
-
Introduction of a leucine half-zipper engenders multiple high-quality crystals of a recalcitrant tRNA synthetase.Acta Crystallogr D Biol Crystallogr. 2010 Mar;66(Pt 3):243-50. doi: 10.1107/S0907444909055462. Epub 2010 Feb 12. Acta Crystallogr D Biol Crystallogr. 2010. PMID: 20179335 Free PMC article.
-
Ferritin Conjugates With Multiple Clickable Amino Acids Encoded by C-Terminal Engineered Pyrrolysyl-tRNA Synthetase.Front Chem. 2021 Nov 25;9:779976. doi: 10.3389/fchem.2021.779976. eCollection 2021. Front Chem. 2021. PMID: 34900939 Free PMC article.
-
Genetic incorporation of histidine derivatives using an engineered pyrrolysyl-tRNA synthetase.ACS Chem Biol. 2014 May 16;9(5):1092-6. doi: 10.1021/cb500032c. Epub 2014 Mar 17. ACS Chem Biol. 2014. PMID: 24506189 Free PMC article.
References
-
- Ibba M, Söll D. Annu Rev Biochem. 2000;69:617–650. - PubMed
-
- Ambrogelly A, Palioura S, Söll D. Nat Chem Biol. 2007;3:29–35. - PubMed
-
- Sauerwald A, Zhu W, Major TA, Roy H, Palioura S, Jahn D, Whitman WB, Yates JR, III, Ibba M, Söll D. Science. 2005;307:1969–1972. - PubMed
-
- Srinivasan G, James CM, Krzycki JA. Science. 2002;296:1459–1462. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

