Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome
- PMID: 17594509
- PMCID: PMC2394776
- DOI: 10.1186/gb-2007-8-6-r127
Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome
Abstract
Background: Transposed elements (TEs) have a substantial impact on mammalian evolution and are involved in numerous genetic diseases. We compared the impact of TEs on the human transcriptome and the mouse transcriptome.
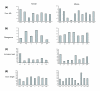
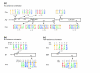
Results: We compiled a dataset of all TEs in the human and mouse genomes, identifying 3,932,058 and 3,122,416 TEs, respectively. We than extracted TEs located within human and mouse genes and, surprisingly, we found that 60% of TEs in both human and mouse are located in intronic sequences, even though introns comprise only 24% of the human genome. All TE families in both human and mouse can exonize. TE families that are shared between human and mouse exhibit the same percentage of TE exonization in the two species, but the exonization level of Alu, a primate-specific retroelement, is significantly greater than that of other TEs within the human genome, leading to a higher level of TE exonization in human than in mouse (1,824 exons compared with 506 exons, respectively). We detected a primate-specific mechanism for intron gain, in which Alu insertion into an exon creates a new intron located in the 3' untranslated region (termed 'intronization'). Finally, the insertion of TEs into the first and last exons of a gene is more frequent in human than in mouse, leading to longer exons in human.
Conclusion: Our findings reveal many effects of TEs on these two transcriptomes. These effects are substantially greater in human than in mouse, which is due to the presence of Alu elements in human.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

