Genetic targeting of the kinase activity of the Met receptor in cancer cells
- PMID: 17595299
- PMCID: PMC2040912
- DOI: 10.1073/pnas.0703205104
Genetic targeting of the kinase activity of the Met receptor in cancer cells
Abstract
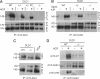
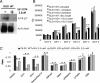
The development of kinase inhibitors is revolutionizing cancer treatment. Assessing the oncogenic potential of individual kinase activities and ensuring that a drug of interest acts by direct inhibition of its putative target kinase are clear priorities. We developed a genetic strategy to selectively inactivate the catalytic activity of kinases. This approach generates isogenic cells in which a given kinase gene is expressed but is devoid of enzymatic activity. As a model to test this approach, we chose the MET receptor, which is involved in multiple cancers and is the focus of several therapeutic efforts. The exon encoding the ATP-binding site of MET was deleted from the genome of colorectal, bladder, and endometrial cancer cells. The derivative isogenic cells expressed a kinase-inactive Met (MET-KD) and were completely unresponsive to its ligand hepatocyte growth factor (HGF), indicating the exclusivity of this ligand-receptor axis. The in vivo tumorigenic potential of MET-KD cells was reduced but could be partially restored by HGF, suggesting that concomitant targeting of the receptor and its ligand should be therapeutically exploited. A reportedly selective Met-kinase inhibitor (SU-11274) markedly affected the growth of MET-KD cancer cells, indicating this compound exerts its effects not only through the intended target. The genetic strategy presented here is not limited to kinase genes but could be broadly applicable to any drug/protein combination in which the target enzymatic domain is known.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
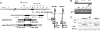

Similar articles
-
The MET receptor tyrosine kinase is a potential novel therapeutic target for head and neck squamous cell carcinoma.Cancer Res. 2009 Apr 1;69(7):3021-31. doi: 10.1158/0008-5472.CAN-08-2881. Epub 2009 Mar 24. Cancer Res. 2009. PMID: 19318576 Free PMC article.
-
Cancer cells harboring MET gene amplification activate alternative signaling pathways to escape MET inhibition but remain sensitive to Hsp90 inhibitors.Cell Cycle. 2009 Jul 1;8(13):2050-6. doi: 10.4161/cc.8.13.8861. Epub 2009 Jul 27. Cell Cycle. 2009. PMID: 19502802 Free PMC article.
-
The flavonols quercetin, kaempferol, and myricetin inhibit hepatocyte growth factor-induced medulloblastoma cell migration.J Nutr. 2009 Apr;139(4):646-52. doi: 10.3945/jn.108.102616. Epub 2009 Feb 25. J Nutr. 2009. PMID: 19244381
-
The function, proteolytic processing, and histopathology of Met in cancer.Adv Cancer Res. 2009;103:1-23. doi: 10.1016/S0065-230X(09)03001-2. Adv Cancer Res. 2009. PMID: 19854350 Review.
-
Isoforms of the met receptor tyrosine kinase.EXS. 1993;65:167-79. EXS. 1993. PMID: 8380736 Review.
Cited by
-
Discordant inheritance of chromosomal and extrachromosomal DNA elements contributes to dynamic disease evolution in glioblastoma.Nat Genet. 2018 May;50(5):708-717. doi: 10.1038/s41588-018-0105-0. Epub 2018 Apr 23. Nat Genet. 2018. PMID: 29686388 Free PMC article.
-
Tivantinib (ARQ 197) affects the apoptotic and proliferative machinery downstream of c-MET: role of Mcl-1, Bcl-xl and Cyclin B1.Oncotarget. 2015 Sep 8;6(26):22167-78. doi: 10.18632/oncotarget.4240. Oncotarget. 2015. PMID: 26259250 Free PMC article.
-
Hepatocyte-specific c-Met deletion disrupts redox homeostasis and sensitizes to Fas-mediated apoptosis.J Biol Chem. 2008 May 23;283(21):14581-9. doi: 10.1074/jbc.M707733200. Epub 2008 Mar 18. J Biol Chem. 2008. PMID: 18348981 Free PMC article.
-
Significance of c-MET overexpression in cytotoxic anticancer drug-resistant small-cell lung cancer cells.Cancer Sci. 2014 Aug;105(8):1032-9. doi: 10.1111/cas.12447. Epub 2014 Jul 25. Cancer Sci. 2014. PMID: 24827412 Free PMC article.
-
Metabolic characterization of colorectal cancer cells harbouring different KRAS mutations in codon 12, 13, 61 and 146 using human SW48 isogenic cell lines.Metabolomics. 2020 Apr 16;16(4):51. doi: 10.1007/s11306-020-01674-2. Metabolomics. 2020. PMID: 32300895 Free PMC article.
References
-
- Bardelli A, Parsons DW, Silliman N, Ptak J, Szabo S, Saha S, Markowitz S, Willson JK, Parmigiani G, Kinzler KW, et al. Science. 2003;300:949. - PubMed
-
- Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, Holcomb T, Pujara K, Stinson J, Fu L, et al. Cancer Res. 2006;66:283–289. - PubMed
-
- Smolen GA, Muir B, Mohapatra G, Barmettler A, Kim WJ, Rivera MN, Haserlat SM, Okimoto RA, Kwak E, Dahiya S, et al. Cancer Res. 2006;66:3452–3455. - PubMed
-
- Jagadeeswaran R, Ma PC, Seiwert TY, Jagadeeswaran S, Zumba O, Nallasura V, Ahmed S, Filiberti R, Paganuzzi M, Puntoni R, et al. Cancer Res. 2006;66:352–361. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

