Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus
- PMID: 17596308
- PMCID: PMC2045430
- DOI: 10.1128/JVI.00771-07
Characterization of the sequence element directing translation reinitiation in RNA of the calicivirus rabbit hemorrhagic disease virus
Abstract
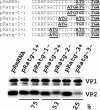
The calicivirus minor capsid protein VP2 is expressed via reinitiation of protein synthesis after termination of translation of the preceding VP1 gene. A sequence element of about 80 nucleotides denoted "termination upstream ribosomal binding site" (TURBS) (25) is crucial for reinitiation. Deletion mapping in the TURBS of a rabbit calicivirus identified two short sequence motifs that were crucial for VP2 expression. Motif 1 is conserved among caliciviruses and is complementary to a sequence in the 18S rRNA. Single-residue exchanges in this motif severely impaired reinitiation when they affected the putative rRNA binding, whereas an exchange preserving complementarity had only a minor effect. Single exchanges in motif 2 were rather well tolerated, but the introduction of double exchanges almost blocked VP2 expression. In contrast, the deletion analyses showed that the RNA between the two motifs is of minor importance. The distance between motif 2 and the start site was found to be important, since deletions of increasing length in this sequence or upstream positioning of the start codon reduced VP2 expression stepwise to low levels, whereas multiple-nucleotide exchanges in this region were tolerated. The low flexibility of the arrangement of TURBS motif 2 and the start codon stand in marked contrast to the requirements with regard to the location of the stop codon of the preceding VP1 gene, which could be moved far downstream with continuous reduction, but without loss, of VP2 translation. The sequence mapping resulted in a refined model of the reinitiation mechanism leading to VP2 expression.
Figures



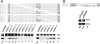



References
-
- Adhin, M. R., and D. J. van Duin. 1990. Scanning model for translational reinitiation in eubacteria. J. Mol. Biol. 213:811-818. - PubMed
-
- Algire, M. A., and J. R. Lorsch. 2006. Where to begin? The mechanism of translation initiation codon selection in eukaryotes. Curr. Opin. Chem. Biol. 10:480-486. - PubMed
-
- Chaudhry, Y., A. Nayak, M. E. Bordeleau, J. Tanaka, J. Pelletier, G. J. Belsham, L. O. Roberts, and I. G. Goodfellow. 2006. Caliciviruses differ in their functional requirements for eIF4F components. J. Biol. Chem. 281:25315-25325. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

