Quantitative analysis of the binding of simian virus 40 large T antigen to DNA
- PMID: 17596312
- PMCID: PMC1951407
- DOI: 10.1128/JVI.00384-07
Quantitative analysis of the binding of simian virus 40 large T antigen to DNA
Abstract
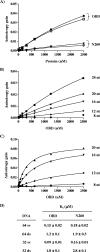
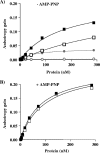
SV40 large T antigen (T-ag) is a multifunctional protein that successively binds to 5'-GAGGC-3' sequences in the viral origin of replication, melts the origin, unwinds DNA ahead of the replication fork, and interacts with host DNA replication factors to promote replication of the simian virus 40 genome. The transition of T-ag from a sequence-specific binding protein to a nonspecific helicase involves its assembly into a double hexamer whose formation is likely dictated by the propensity of T-ag to oligomerize and its relative affinities for the origin as well as for nonspecific double- and single-stranded DNA. In this study, we used a sensitive assay based on fluorescence anisotropy to measure the affinities of wild-type and mutant forms of the T-ag origin-binding domain (OBD), and of a larger fragment containing the N-terminal domain (N260), for different DNA substrates. We report that the N-terminal domain does not contribute to binding affinity but reduces the propensity of the OBD to self-associate. We found that the OBD binds with different affinities to its four sites in the origin and determined a consensus binding site by systematic mutagenesis of the 5'-GAGGC-3' sequence and of the residue downstream of it, which also contributes to affinity. Interestingly, the OBD also binds to single-stranded DNA with an approximately 10-fold higher affinity than to nonspecific duplex DNA and in a mutually exclusive manner. Finally, we provide evidence that the sequence specificity of full-length T-ag is lower than that of the OBD. These results provide a quantitative basis onto which to anchor our understanding of the interaction of T-ag with the origin and its assembly into a double hexamer.
Figures






References
-
- Alexandrov, A. I., M. R. Botchan, and N. R. Cozzarelli. 2002. Characterization of simian virus 40 T-antigen double hexamers bound to a replication fork. The active form of the helicase. J. Biol. Chem. 277:44886-44897. - PubMed
-
- Baur, C. P., K. Klausing, M. Scheffner, H. Stahl, and R. Knippers. 1988. Protein-DNA interactions at the simian virus 40 origin of replication. Biochim. Biophys. Acta 951:388-395. - PubMed
-
- Borowiec, J. A., F. B. Dean, P. A. Bullock, and J. Hurwitz. 1990. Binding and unwinding—how T antigen engages the SV40 origin of DNA replication. Cell 60:181-184. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

