In vivo and in vitro intragenomic rearrangement of TT viruses
- PMID: 17596318
- PMCID: PMC1951432
- DOI: 10.1128/JVI.00781-07
In vivo and in vitro intragenomic rearrangement of TT viruses
Abstract
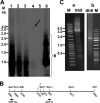
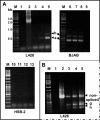
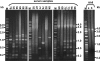
The in vitro replication of the Torque teno virus (TT virus) tth8 full-length genome and particle formation in a Hodgkin's lymphoma-derived cell line after transfection with cloned viral DNA were demonstrated. Analyses of the transcription patterns of tth8 and tth7 TT virus isolates in a number of lymphoma and T-cell leukemia cell lines indicated differential additional splicing events and intragenomic rearrangement generating open reading frames which could not be deducted from the genomic sequence. We also demonstrated the presence of rearranged TT virus genomes in vivo in sera taken from pregnant mothers whose children later developed childhood leukemia, as well as sera from control mothers. Control experiments using religated cloned genomic tth8 DNA mixed with cellular DNA did not result in such subviral molecules. These subviral isolates ranged from 172 bp to full-length TT virus genomes. Possible in vivo selection for specific rearranged molecules was indicated by the presence of one isolate (561 bp) in 11 serum samples. It remains to be clarified whether selected rearranged subviral components resulting from specific TT virus types may contribute to the initiation of disease. These data demonstrate new features of TT viruses suggesting possible similarities to plant viruses of the family Geminiviridae, as well as raise questions about the documented plurality and diversity of anelloviruses.
Figures


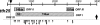

References
-
- Aebi, M., H. Hornig, R. A. Padgett, J. Reiser, and C. Weissmann. 1986. Sequence requirements for splicing of higher eukaryotic nuclear pre-mRNA. Cell 47:555-565. - PubMed
-
- Amin, I., S. Mansoor, L. Amrao, M. Hussain, S. Irum, Y. Zafar, S. E. Bull, and R. W. Briddon. 2006. Mobilization into cotton and spread of a recombinant cotton leaf curl disease satellite. Arch. Virol. 151:2055-2065. - PubMed
-
- Ball, J. K., R. Curran, S. Berridge, A. M. Grabowska, C. L. Jameson, B. J. Thomson, W. L. Irving, and P. M. Sharp. 1999. TT virus sequence heterogeneity in vivo: evidence for co-infection with multiple genetic types. J. Gen. Virol. 80:1759-1768. - PubMed
-
- Biagini, P., P. Gallian, H. Attoui, M. Touinssi, J.-F. Cantaloube, P. de Micco, and X. de Lamballerie. 2001. Genetic analysis of full-length genomes and subgenomic sequences of TT virus-like mini virus human isolates. J. Gen. Virol. 82:379-383. - PubMed
-
- Cacoub, P., E. Rosenthal, V. Gerolami, P. Hausfater, P. Ghillani, Y. Sterkers, V. Thibault, H. Khiri, J. C. Piette, and P. Halfon. 2003. Transfusion-associated TT virus co-infection in patients with hepatitis C virus is associated with type II mixed cryoglobulinemia but not with B-cell non-Hodgkin lymphoma. Clin. Microbiol. Infect. 9:39-44. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

