Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import
- PMID: 17596514
- PMCID: PMC1951752
- DOI: 10.1091/mbc.e07-01-0088
Multiple 40-kDa heat-shock protein chaperones function in Tom70-dependent mitochondrial import
Abstract
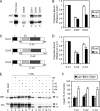
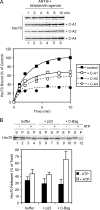
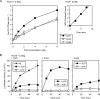
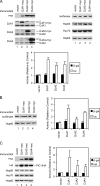
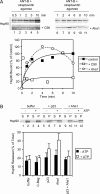
Mitochondrial preproteins that are imported via the translocase of the mitochondrial outer membrane (Tom)70 receptor are complexed with cytosolic chaperones before targeting to the mitochondrial outer membrane. The adenine nucleotide transporter (ANT) follows this pathway, and its purified mature form is identical to the preprotein. Purified ANT was reconstituted with chaperones in reticulocyte lysate, and bound proteins were identified by mass spectrometry. In addition to 70-kDa heat-shock cognate protein (Hsc70) and 90-kDa heat-shock protein (Hsp90), a specific subset of cochaperones were found, but no mitochondria-specific targeting factors were found. Interestingly, three different Hsp40-related J-domain proteins were identified: DJA1, DJA2, and DJA4. The DJAs bound preproteins to different extents through their C-terminal regions. DJA dominant-negative mutants lacking the N-terminal J-domains impaired mitochondrial import. The mutants blocked the binding of Hsc70 to preprotein, but with varying efficiency. The DJAs also showed significant differences in activation of the Hsc70 ATPase and Hsc70-dependent protein refolding. In HeLa cells, the DJAs increased new protein folding and mitochondrial import, although to different extents. No single DJA was superior to the others in all aspects, but each had a profile of partial specialization. The Hsp90 cochaperones p23 and Aha1 also regulated Hsp90-preprotein interactions. We suggest that multiple cochaperones with similar yet partially specialized properties cooperate in optimal chaperone-preprotein complexes.
Figures




References
-
- Abe Y., Shodai T., Muto T., Mihara K., Torii H., Nishikawa S., Endo T., Kohda D. Structural basis of presequence recognition by the mitochondrial protein import receptor Tom20. Cell. 2000;100:551–560. - PubMed
-
- Borges J. C., Fischer H., Craievich A. F., Ramos C. H. Low resolution structural study of two human HSP40 chaperones in solution. DJA1 from subfamily A and DJB4 from subfamily B have different quaternary structures. J. Biol. Chem. 2005;280:13671–13681. - PubMed
-
- Bose S., Weikl T., Bugl H., Buchner J. Chaperone function of Hsp90-associated proteins. Science. 1996;274:1715–1717. - PubMed
-
- Brix J., Ziegler G. A., Dietmeier K., Schneider-Mergener J., Schulz G. E., Pfanner N. The mitochondrial import receptor Tom 70, identification of a 25 kDa core domain with a specific binding site for preproteins. J. Mol. Biol. 2000;303:479–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

